The novel coronavirus SARS-CoV-2 (COVID-19) is a member of the family Coronaviridae and contains ssRNA genome. The emergency of COVID-19 has caused global threatened and panic health security. In order to detect common regions and genes of the Severe acute respiratory syndrome-associated coronavirus 2, we collected the whole genome of all viruses available in databases for this family (55 complete genomes), and made comparative genomic analyses with the collected data. We performed an interactomics approach to identify miRNAs that could be affected in some regions of the whole virus genome. As well as, protein structure modeling was used for modeling of related sequence. Cladogram revealed Bat coronavirus, MERS-related coronavirus, SARS-related coronavirus and SARS coronavirus 2 are closely related. The most important genes involved in the disease were RELA in virus genome and
ACE
2 receptors and
CLEC
4M genes in the host genome. RELA gene was suppressed by hsa-miR-516b-3p, hsa-miR-3529-3p and hsa-miR-6749-3p, ACE2 receptor was suppressed by hsa-miR-23b-5p and hsa-miR-769-5p, and finally, hsa-miR-4462 and hsa-miR-5187-5p suppressed CLEC4M gene. Therefore, our results will help to control and treat COVID-19 and revealed new insight into the vaccine design and miRNA therapy.
COVID-19, comparative genomics, miRNA, miRNA therapy, interactomics
The outbreak of person-to-person transmissible and atypical caused by the COVID-19 has caused global concern. There have been more than 1,200,000 approved cases of this disease in the world, as of April 7, 2020. According to the World Health Organization, 16-21% of people with the virus have become ill with a 2-3% mortality rate [1]. Therefore, it is crucial to identify an effective method as early as possible for vaccine design and treatment procedures.
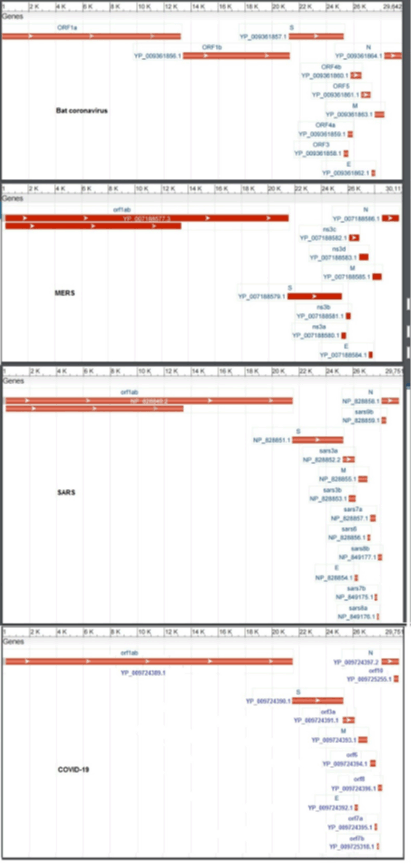
Although some studies have reported clinical characteristics of SARS, MERS and SARS2 [2-5] genomic comparison of complete genome features and suggested treatments for COVID-19 has not been reported yet. The Severe acute respiratory syndrome coronavirus 2 is the seventh member of enveloped RNA coronaviruses [6-8]. A typical coronaviruses family contains four main structural proteins: S (spike protein), M (membrane protein), E (envelope protein), and N (nucleocapsid protein), respectively [9].
Besides, the interactomics approach and miRNA-gene analyses have mostly been used as complementary approaches for extracting biological information from omics layers and increased awareness of the complex disease [10].
Most studies considered neither non-coding region nor the detection of genomic regions associated with COVID-19, which is an important aspect of vaccine design and treatment patients. Therefore, this study aims to demonstrate how new genomic scale comparison and new approaches can provide new insights into the procedures that govern the emergence and evolution of coronaviruses and how to treat related diseases. In doing, so we make general statements about the nature of ssRNA virus evolution and highlight some of the key evolutionary mechanisms. As a sidebar, this work showed the increasingly important role played by comparative genomics and applying the interactomics approach in the study of COVID-19.
All of the used software, databases, online/offline tools are listed in Table 1.
Table 1. Software, databases, online/offline tools, generally explanations and website addresses
Software name |
Explanations |
Accession web Address |
Viral genome browser |
The viral genome browser includes 9240 complete genomes. The header part lists taxonomy groups. The genome table lists species and reference genomes (RefSeqs) within a selected taxonomy group. |
https://www.ncbi.nlm.nih.gov/genome/viruses |
Gene Expression Omnibus |
GEO is a public functional genomics data repository. Tools are provided to help users query and download experiments and curated gene expression profiles. |
https://www.ncbi.nlm.nih.gov/geo |
Functional Annotation Tools |
Gene annotation involves the process of taking the raw DNA sequence produced by the genome-sequencing projects and adding layers of analysis and interpretation necessary to extracting biologically significant information and placing such derived details into context. |
https://david.ncifcrf.gov |
Multiple Sequence Alignment (MSA) |
MSA is generally the alignment of three or more biological sequences (protein or nucleic acid) of similar length. From the output, homology can be inferred and the evolutionary relationships between the sequences studied. |
www.ebi.ac.uk/Tools/msa |
Phylogenetic Tree |
This tool provides access to phylogenetic tree generation methods from the ClustalW2 package. |
www.ebi.ac.uk/Tools/phylogeny |
CoGe |
CoGe is a platform for performing Comparative Genomics research. It provides an open-ended network of interconnected tools to manage, analyze, and visualize next-gen data. |
genomevolution.org/coge |
BLASTn,p,x |
The Basic Local Alignment Search Tool (BLAST) finds regions of local similarity between sequences. |
blast.ncbi.nlm.nih.gov/Blast.cgi |
Protein modeling |
The aim of the SWISS-MODEL Repository is to provide access to an up-to-date collection of annotated 3D protein models generated by automated homology modelling for relevant model organisms and experimental structure information for all sequences in UniProtKB. |
swissmodel.expasy.org/interactive#sequence |
HHPred |
Homology detection and structure prediction by HMM-HMM comparison |
toolkit.tuebingen.mpg.de/tools/hhpred |
Literature Mining |
It allows researchers to explore a network of gene and protein interactions based on published scientific literature. |
www.ihop-net.org/UniPub/iHOP |
miRWalk 2.0 and 3.0 |
The platform integrates information from different miRNA-target databases, including validated information and prediction datasets: MiRWalk, miRDB, miRMap, miRNAMap, MicroT4, miRanda, miRBridge, PICTAR2, RNAhybrid, Targetscan, PITA and RNA22. |
zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/index.html |
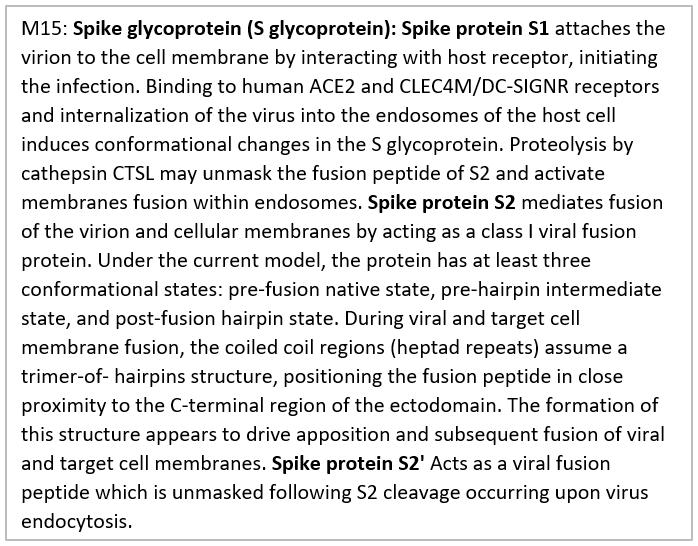
Structure and comparative genomics of the family Coronaviridae complete genomes were performed based on related complete genome. For this purpose, all published data related to the whole-genome of viruses were collected; a total of 55 complete genomes were used for the next step of the analysis. One of the most important goals of comparative genomics is finding differences between various strains and the use of these differences for the next goals such as the identification of genes and miRNAs. We have identified a total of 459 miRNAs that suppress RELA, ACE2 receptor and CLEC4M genes but we have only reported gene-specific miRNAs and this study one of the first studies in the field of molecular therapeutic specially treatment of COVID-19. miRNAs that suppress these genes were listed as supplementary Table 1. We have performed based on virus RNA sequences and observed high homology (>99%) between nsp1 or leader protein with RELA or p56 gene, as well as, protein homology detection and modeling confirmed mentioned gene. Also, ACE2 receptor and CLEC4M genes were related to S protein or Spike glycoprotein that spike protein S1 attaches the virion to the cell membrane by interacting with host receptor, initiating the infection. Binding to human ACE2 receptor and CLEC4M genes and internalization of the virus into the endosomes of the host cell induces conformational changes in the S glycoprotein. Proteolysis by cathepsin CTSL may unmask the fusion peptide of S2 and activate membranes fusion within endosomes. Spike protein S2 mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein (www.uniprot.org) (supplementary Table 2).
The miRNAs pair with genes or mRNAs to induce gene or mRNA translational repression and degradation [11]. The various molecular procedures at the mRNA destabilization and heart of miRNA-directed translational repression, that include inhibition of translation initiation and poly(A) shortening, are reviewed elsewhere [12]. Within a region of the 3'-UTR (3'-untranslated region) of the RELA, ACE2 receptor and CLEC4M genes in COVID-19 disease were suppressed by mentioned miRNAs.
miRNAs are critical for normal animal growth and are involved in a difference of biological pathways [13]. Expression of miRNAs is associated with many human diseases [14,15]. As well as, miRNAs are secreted into extracellular fluids. Extracellular miRNAs have been widely reported as potential biomarkers for different biological process and they also serve as signaling molecules to mediate cell-cell communications [16].
Phylogenetic analyses of complete genome sequence data have revealed that Bat coronavirus, Middle East respiratory syndrome-related coronavirus, Severe acute respiratory syndrome-related coronavirus and Severe acute respiratory syndrome coronavirus 2 were closely related (Figure 1). This comparison dedicates that the origin of the virus could be held in common and the same strategies can be used to treat them. In addition, by Focusing on the genome structure of these four strains revealed little differences. The most similarity was between the Severe acute respiratory syndrome-related coronavirus and Severe acute respiratory syndrome coronavirus 2 genomes (Supplementary Figure 1). Multiple sequences alignment (MSA) also confirmed this statement (Figure 2).
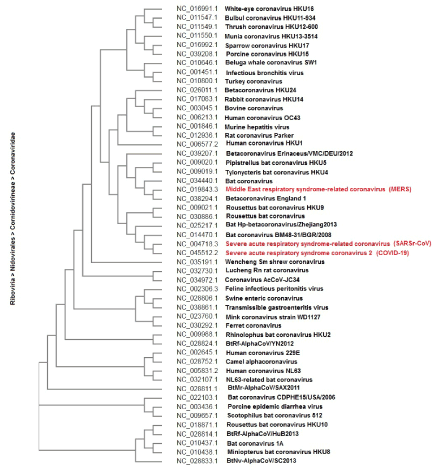
Figure 1. Cladogramof the family Coronaviridae complete genomes were reconstructed by Clustal Omega using 55 sequences with the best fitting evolutionary model. Subsequently, the tree was purged from the most similar sequences and midpoint rooted. GenBank accession numbers for all viruses are shown.
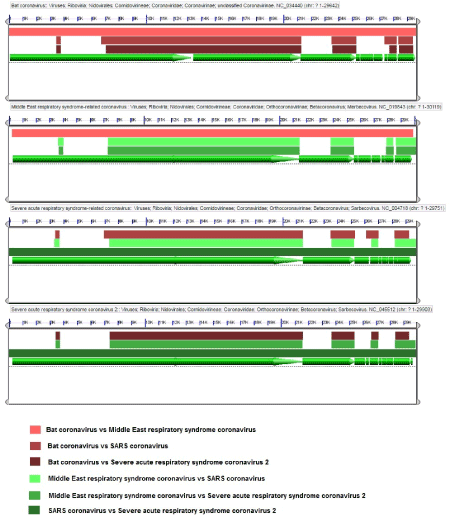
Figure 2. Multiple sequence alignments (MSAs) of the sequenses combination using Bat coronavirus, Middle East respiratory syndrome-related coronavirus, Severe acute respiratory syndrome-related coronavirus and Severe acute respiratory syndrome coronavirus 2 by LASTZ
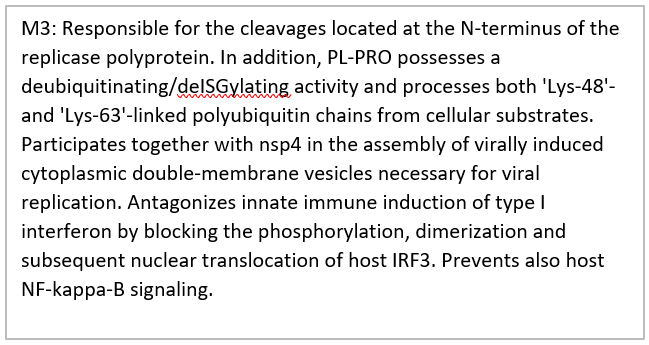
Another important aspect of comparative genomics is the comparison of mutation rates and changes in protein structure. So, we studied the protein structure changes, and also evaluated the homology between the different parts of the virus protein with the relevant databases (Figure 3). Protein orf1ab divided into 18 parts that involved: bases 266-805 encode nsp1 and produce leader protein. This protein has high homology with p65 protein. Transcription factor p65 also known as nuclear factor NF-kappa-B p65 subunit is a protein that in humans is encoded by the RELA gene. Gene expression regulation by RELA is fundamental for controlling many important procedures, including immune responses, apoptosis, inflammatory, cell proliferation and development [17]. Up-regulation of this gene related to many cancers and activation has been found to be correlated with cancer development [18]. Using miRNAs that suppressed this gene can disrupt the replication of the virus. In this regard, we identified three miRNAs that increasing the blood concentration of these miRNAs that can suppress the activity of the RELA gene. hsa-miR-516b-3p, hsa-miR-3529-3p and hsa-miR-6749-3p suppressed mentioned gene. Therefore, could be used as a drug that paralyzing for the virus. Bases 806-2719 encode nsp2 and this protein may play a key role in the modulation of host cell survival signaling pathway by interacting with host PHB1 and PHB2. PHB1 and PHB2 belong to the prohibition domain family, and both exist in different cellular compartments such as cell membrane, nucleus and the mitochondria. Many studies have reported differential expression of the PHB1 and PHB2 in cancers. Furthermore, studies confirmed that PHB1 and PHB2 are involved in the biological procedures of tumorigenesis, including metastasis, apoptosis and cell proliferation [19]. Bases 2720-8554, 2720-8554, 2720-8554, 2720-8554 and 2720-8554 that encode nsp3, ADP-ribose-1"-monophosphatase, Replicase polyprotein 1ab, nsp3 and Replicase polyprotein 1ab, respectively. These regions are responsible for the cleavages located at the N-terminus of the replicase polyprotein. In addition, it participates together with nsp4 in the assembly of virally induced cytoplasmic double-membrane vesicles necessary for viral replication. Bases 8555-10054 encode nsp4 and produce RNA-directed RNA polymerase.
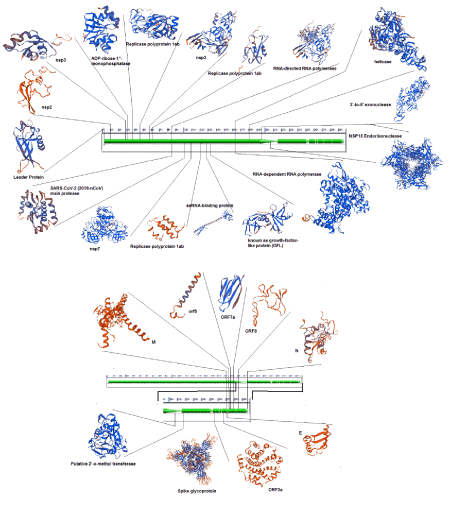
Figure 3. Protein structure modeling for Severe acute respiratory syndrome coronavirus 2 by ExPASy tools
Bases 10055-10972 encode SARS-CoV-2 (2019-nCoV) main protease and produce 3C-like proteinase. Bases 11843-12091, 12092-12685 and 12686-13024 encode nsp7, Replicase polyprotein 1ab and ssRNA-binding protein, respectively. Bases 13025-13441encode nsp10 that known as growth-factor-like protein (GFL). This part plays an essential role in viral mRNAs cap methylation. Bases 13025-13441 encode nsp12 and produce RNA-dependent RNA polymerase. Bases 16237-18039 encode nsp13 and produce helicase. Bases 18040-19620 encode 3'-to-5' exonuclease. 3'-to-5' exonuclease processing two different activities: an N7-guanine methyltransferase activity and an exoribonuclease activity acting on both dsRNA and ssRNA. Bases 19621-20658 encode NSP15 Endoribonuclease (Mn(2+)-dependent, uridylate-specific enzyme). Bases 20659-21552 encode putative 2'-o-methyl transferase. Bases 21563-25384 that encode spike glycoprotein, this protein attach the virion to the cell membrane by interacting with the host receptor, initiating the infection.
The finding that SARS-2-S exploits ACE2 for entry, which was also reported [20]. However, ACE2 expression is not limited to the lung, and extrapulmonary spread of SARS-CoV in ACE2+ tissues were reported [21]. Thus, increasing the blood concentration of these miRNAs can suppress the activity of these genes and prevented the virus from progressing. ACE2 receptor was suppressed by hsa-miR-23b-5p and hsa-miR-769-5p, and, hsa-miR-4462 and hsa-miR-5187-5p suppressed CLEC4M gene. Bases 25393-26220, 26245-26472, 26523-27191, 27202-27387, 27394-27759, 27756-27887 and 27894-28259 that encode ORF3a, E, M, orf6, ORF7a, ORF7b and ORF8 proteins, respectively. Bases 28274-29533 encode N protein and produce nucleocapsid phosphoprotein, and finally, bases 29558-29674 encode ORF10 protein. Therefore, the most important genes are RELA, ACE2 receptor and CLEC4M gene. By suppressing these genes, can hope to get treatment for this disease. One of the strategies used to treat cancer is to utilize non-coding RNAs such as miRNAs. In other words, systems biology and the interactomics approach are a systemic level approach to study an all-around understanding of complicated biological systems outside the molecular-level scale [22]. Instead of analyzing individual components or aspects of the organism, such as a cell nucleus or metabolism, systems biologists focus on all aspects and the interactions between them as part of one system such as using the gene-miRNA approach. Therefore, this method of considering miRNA–gene interactome will help to clarify the complicated biological procedures for this disease. microRNAs are small non-coding RNAs that regulate gene expression post-transcriptionally by interfering with the translation of one or more target genes. The dysregulation of miRNAs contributes to the pathogenesis of all types of cancer.
The recent decade has witnessed a substantial improvement of miRNA replacement therapy. This approach aims to restore disease suppressor miRNA function in some cells using miRNA expression plasmids or synthetic miRNA mimics. Recent advances in miRNA replacement therapy for the treatment of cancer and its advantages were reported [23]. The fact that various miRNA replacement therapies are currently in the clinical trial shows the great potential of this approach to treat disease. In the same direction, we applied miRNA databases for identifying miRNAs that suppress motioned genes. We believe that the mentioned approach could treat COVID-19, however, further experimental tests are needed. Therefore, this study could enhance the identification of biological mechanisms and respective candidate genes, and also, useful in disease therapy.
Data availability statement
All datasets generated for this study are included in the article/supplementary material.
Ethics statement
Ethical review and approval were not required for the study on human participants in accordance with the local legislation and institutional requirements.
Declaration of interests
The author declares that they have no competing interests.
Funding
Not applicable.
Acknowledgements
I would like to thank Prof. Reza Miraie-Ashtiani and Prof. Mostafa Sadeghi for providing critical feedback and reagents, Fatemeh Marandi for technical assistance and the University of Tehran, Department of Animal Science. I also thank the anonymous editor and reviewers for their constructive feedback which significantly improved the quality of our analysis.
View Supplementary Data
- Yang Y, Lu Q, Liu M, Wang Y, Zhang A, et al. (2020) Epidemiological and clinical features of the 2019 novel coronavirus outbreak in China. medRxiv.
- Huang C, Wang Y, Li X, Ren L, Zhao J, et al. (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395: 497-506.
- Wang D, Hu B, Hu C, Zhu F, Liu X, et al. (2020) Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA 323: 1061-1069.
- Ren J, Zhang AH, Wang XJ (2020) Traditional Chinese medicine for COVID-19 treatment. Pharmacol Res 10: 47-43.
- Al-Baadani AM, Elzein FE, Alhemyadi SA, Khan OA, Albenmousa AH, et al. (2019) Characteristics and outcome of viral pneumonia caused by influenza and Middle East respiratory syndrome-coronavirus infections: A 4-year experience from a tertiary care center. Ann Thorac Med 14: 179-185. [Crossref]
- Peiris JS, Guan Y, Yuen KY (2004) Severe acute respiratory syndrome. Nat Med 10: 88-97.
- Lee J, Chowell G, Jung E (2016) A dynamic compartmental model for the Middle East respiratory syndrome outbreak in the Republic of Korea: A retrospective analysis on control interventions and superspreading events. J Theor Biol 408: 118-126.
- Zhu N, Zhang D, Wang W, Li X, Yang B, et al. (2020) A novel coronavirus from patients with pneumonia in China. N Engl J Med 382: 727-733.
- Grifoni C, Sidney J, Zhang Y, Scheuermann RH, Peters B, et al. (2020) A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host & Microbe 27: 671-680.
- Bahrami A, Miraie-Ashtiani SR, Sadeghi M, Najafi A (2017) miRNA-mRNA network involved in folliculogenesis interactome: Systems biology approach. Reproduction 154: 51-65. [Crossref]
- O'Brien J, Hayder H, Zayed Y, Peng C (2018) Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol 9: 402. [Crossref]
- Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102-114.
- Fu G, Brkic J, Hayder H, Peng C (2013) MicroRNAs in human placental development and pregnancy complications. Int J Mol Sci 14: 5519-5544.
- Tufekci KU, Oner MG, Meuwissen RL, Genc S (2014) The role of microRNAs in human diseases. Methods Mol Biol 110: 33-50.
- Paul P, Chakraborty A, Sarkar D, Langthasa M, Rahman M, et al. (2018) Interplay between miRNAs and human diseases. J Cell Physiol 233: 2007-2018. [Crossref]
- Huang W (2017) MicroRNAs: biomarkers, diagnostics, and therapeutics. Methods Mol Biol 1617: 57-67. [Crossref]
- Hayden MS, West AP, Ghosh S (2006) NF-_B and the immune response. Oncogene 25: 6758-6780.
- Vlahopoulos S, Adamaki M, Khoury N, Zoumpourlis V, Boldogh I (2019) Roles of DNA repair enzyme OGG1 in innate immunity and its significance for lung cancer. Pharm Therap 194: 59-72. [Crossref]
- Desideri E, Cavallo AL, Baccarini M (2015) Alike but different: RAF paralogs and their signaling outputs. Cell 161: 967-970. [Crossref]
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, et al. (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579: 270-273. [Crossref]
- Gu X (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan. Lancet.
- Najafi A, Bidkhori G, Bozorgmehr JH, Koch I, Masoudi-Nejad A (2014) Genome scale modeling in systems biology: algorithms and resources. Curr Genom 15: 130-159. [Crossref]
- Hosseinahli N, Aghapour M, Duijf PHG, Baradaran B (2018) Treating cancer with microRNA replacement therapy: A literature review. J Cell Physiol 233: 5574-5588. [Crossref]