Abstract
Morphogenesis is related to Candida albicans pathogenesis and nutritional/ environmental factors linked to eIF2-mediated translational regulation, modulated by Gcn2 and Gcn4, are common inducers of morphological changes. Here we show that C. albicans GCN2 knockout is less virulent to Caenorhabditis elegans and worms survive longer (p < 0.05) than control worms infected with wild-type yeast. Besides, C. albicans GCN4 knockout showed slightly reduced virulence to C. elegans when compared to controls (p <0.05). Interestingly, C. elegans decreased susceptibility to C. albicans GCN2 knockout correlates with its decreased filamentous growth and biofilm formation ability in amino acid rich media used to survival assays (p <0.001). We conclude that Gcn2 kinase, regulator of General Amino Acid control pathway (GAAC) and its target Gcn4 are important to C. albicans virulence in C. elegans model of infection.
Key words
GCN2, GCN4, eIF2, Candida albicans
Introduction
Morphological changes related to microbial pathogenesis are commonly referred as virulence factors and are observed in some pathogenic fungi including the genera Candida that cause a frequent diagnosed human fungal infection [1,2]. Similar to other dimorphic pathogenic fungi, Candida albicans can differentiate from yeast to micelial form and vice versa and this dimorphism is central to biofilm formation, infection and disease [3-6].
Ambient temperatures >35°C, serum, neutral pH, nutrient starvation, N-acetylglucosamine and CO2 promote yeast to hypha morphogenesis in C. albicans [7,8]. Morphogenesis in C. albicans is also regulated by quorum sensing [9] and MAPK/ Ras-cAMP signaling pathways in response to starvation and/or serum signals [10-13]. In addition, previous work reported that besides their role as nutritional stress sensors, eIF2 kinase Gcn2 and its indirect target Gcn4 participate in C. albicans morphogenesis during amino acid starvation [14,15]. Here we report that Gcn2 coordinates the virulence of C. albicans in the well-established C. elegans model of infection [16-18] and is related to morphological switch between yeast and micelial forms, as well as involved in biofilm formation during amino acid rich growth conditions.
Materials and methods
Strains and media
The C. albicans strain CAI-4 (ura3::l imm434/ura3::l imm434), C. albicans Dgcn2 (like CAI-4 except, gcn2::hisG/gcn2::hisG) [14], C. albicans Dgcn4 (like CAI-4 except, gcn4::hisG/gcn4::hisG) [14] were maintained on yeast extract peptone dextrose agar (YPD - Difco). The standard C. elegans strain N2 Bristol was maintained at 15°C and propagated on E. coli strain OP50 using established procedures [19,20].
C. elegans survival assays
Yeast lawns were grown for survival assays as follows: yeast strains were inoculated into 2 ml of YPD and grown at 30°C for 18 h; 30 ml of the culture was spread on 35-mm tissue-culture plates (Falcon) containing nematode growth media (NGM), [19]. The plates were incubated at 30°C overnight and then for two days at 25°C. At the experiment day, a Pasteur pipette molded into the shape of hockey stick was used to gently scrape excess of yeast of the top of the thick yeast lawn. This step facilitated scoring the animals as live or dead on subsequent days. For all experiments Ampicillin (100 mg/ml - Sigma) was added to the medium to selectively prevent growth of E. coli OP50 carried over on transfer of worms to the yeast-containing plates. Between 30 and 40 C. elegans animals at the young adult developmental stage (L4) were transferred from a lawn of E. coli OP50 on NGM to a lawn of the yeast to be tested on NGM media, incubated at 25°C, and examined for viability at 24h intervals with a dissecting microscope. Worms were considered dead when they did not respond to touch with a platinum wire pick. Each experimental condition was tested in triplicate. Plotting of killing curves, calculation of LT50 (time for half of the worms to die) and estimation of differences in survival (log-rank and Wilcoxon tests) with the Kaplan–Meier method were performed by using STATA 6 statistical software (Stata, College Station, TX). P values < 0.05 were considered significant.
Phenotypic analysis of C. albicans knockouts
To evaluate potential morphologic changes of C. albicans during C. elegans survival assays, one isolated colony of wild type and knockouts (Dgcn2 or Dgcn4), growth in YPD plates at 30°C, were inoculated in fresh liquid YPD and incubated 18 hours at 30°C. The cultures were diluted (1:10) in YPD, incubated until reach O.D.600 = 0.3 and then spotted on NGM plates and YPD plates. Morphological changes were evaluated by macroscopic and microscopic observation of spots after incubation at 25°C and 30°C for 72 hours.
Biofilm formation assay
To prepare a standard cell suspension, a single colony of wild type and knockout C. albicans strains was cultured in Synthetic Complete medium (SC) and incubated for 18 h at 30°C with agitation. The fungal cells were harvested by centrifugation, washed twice in PBS (pH 7.2), and resuspended at 1×107 cells/ml. RPMI-1640 media (Invitrogen) and liquid NGM media were used for hyphal induction at 37°C. Fungal biofilms were prepared and analyzed as previously described [21] on commercially available, pre-sterilized, flat-bottomed 96-well polystyrene microtitre plates (Corning). Standard yeast cell suspension (100 µl) was transferred into the plate wells and incubated for 24h at 37°C with agitation. After adhesion phase, the liquid was aspirated, and each well was washed twice with PBS to remove loosely attached cells. The metabolic activity of the C. albicans biofilms was determined quantitatively using a standard 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction assay. The data were analyzed employing a two-way ANOVA and Bonferroni post-test.
Results
Survival assays of C. elegans infected with C. albicans
To verify if GCN2 and GCN4 are important to yeast virulence, we performed C. elegans survival assays with C. albicans WT (CAI-4), Dgcn2 or Dgcn4. As shown in figure 1A, C. elegans infected with C. albicans Dgcn2 is significantly more resistant to fungal damages (LT50 = 5 days, time required to kill 50% of worms; p <0.05) than worms infected with wild type C. albicans (LT50=3 days). Deletion of GCN4, the indirect target of Gcn2, in C. albicans showed decrease in nematode survival after infection at the first days of our assays (LT50= 2 days; p <0.05) when compared to WT infection (LT50= 3 days). Despite the survival assay results, C. elegans showed decreased susceptibility to C. albicans Dgcn4 at the end of the experiment (Figure 1B), since worms stop to die 3 days post-infection.
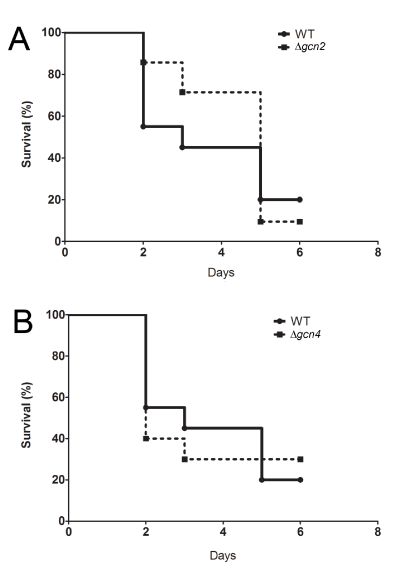
Figure 1. GCN2 and GCN4 are essential for C. albicans virulence in C. elegans.
A) C. albicans Dgcn2 showing significant reduction of virulence (p <0.05) to C. elegans (LT50 = 5 days) when compared to control yeast (wild-type – WT, CAI-4) (LT50 = 3 days). B) Survival of C. elegans after infection with wild type (LT50=3 days) or C. albicans Dgcn4 (LT50 = 2 days) showing worms resistance 3 days after infection when compared to WT.
Phenotypic analysis of C. albicans knockout strains in NGM culture medium
We performed macroscopic and microscopic analysis of C. albicans Dgcn2 and Dgcn4 grown on YPD and NGM agar to verify potential changes in growth patterns. We did not observe differences in the growth rate of C. albicans WT and knockouts spotted on YPD agar (data not shown). As observed in figure 2, we demonstrate that NGM media is permissive to C. albicans morphologic transition at 30°C and at the temperature used in C. elegans survival assays (25°C). At analyzed temperatures C. albicans WT showed differentiation from yeast to filamentous form (Figure 2A, B and C). Interestingly, C. albicans Dgcn4 showed the same morphologic changes observed in wild type strain but with fewer filaments (Figure 2D, E and F). On the other side, C. albicans Dgcn2 showed decreased growth capacity and complete absence of filamentous growth in NGM (Figure 2G, H and I).
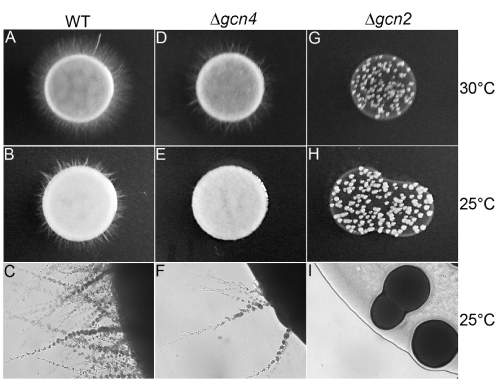
Figure 2. Virulence of C. albicans Dgcn2 and Dgcn4 to C. elegans is related to hyphae production in Nematode Growth Media. C. albicans WT, Dgcn2 and Dgcn4 were grown in NGM agar plates at 30°C and 25°C, as indicated. Wild-type C. albicans changed to its filamentous form at both tested temperatures (A, B and C). GCN4 (Dgcn4) knockout showed reduced morphological transition at 30°C and 25°C when compared to WT strain (D, E and F); GCN2 (Dgcn2) knockout showed decreased growth rate and absence of morphological transition to hyphae on NGM (G,H and I). A, B, D, E, G and H, yeast spot photo. C, F and I, 10X magnification.
C. albicans GCN2 knockout biofilm formation assay
In order to evaluate if absence of filamentous growth in C. albicans GCN2 knockout observed in NGM media correlates with biofilm formation ability, we performed a biofilm formation assay using MTT reduction assay as maker of yeast proliferative activity. We found that the C. albicans Dgcn2 have significantly reduced capacity to form biofilm in RMPI-1640 and NGM media (p <0.001) when compared to wild-type yeast at 25°C (data not shown) and 30°C (Figure 3).
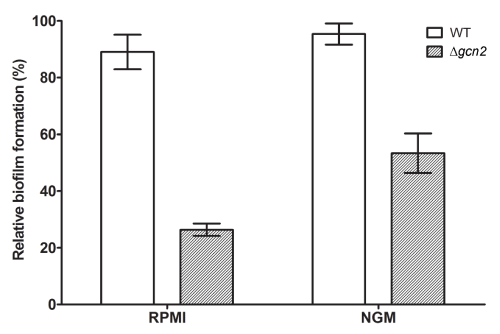
Figure 3. C. albicans Dgcn2 is defective in biofilm formation. Biofilm formation of C. albicans Dgcn2 was examined in RPMI-1640 and NGM medium (30°C). The biofilm metabolic activity was assessed quantitatively using MTT reduction assay. The activity of wild type C. albicans was taken as 100%. Results shown were the average of three independent experiments ± SD. p <0.0001 when compared with WT yeast.
Discussion
Inhibition of global protein synthesis is a main pathway driving stress responses in eukaryotes resulting in remodeling of gene expression patterns, changing the proteome and maintaining homeostasis [22,23]. In yeast, translation can be regulated by eIF2-mediated mechanisms involving only one conserved eIF2 kinase (Gcn2) that phosphorylates the alpha subunit of eIF2, shutting-off global protein synthesis and promoting the translation of mRNA encoding the transcriptional regulator Gcn4 [22].
Gcn2 have important roles on the morphogenesis and virulence of protozoa regulating adaptation of proliferative forms to host and enabling parasite to overcome exposure to the host extracellular environment [24-27]. There is limited information on the role of Gcn2 and Gcn4 in adaptation and virulence of fungal pathogens to their hosts. In vitro analysis showed that Gcn2 and Gcn4 are involved in C. albicans morphogenesis induced by N-acetylglucosamine (GlcNAc), suggesting relevant role of Gcn2 during C. albicans pathogenesis [28]. Here we show that eIF2-mediated translation regulation, besides its function during adaptation to nutritional and environmental stress [14,29,30], is also essential to yeast virulence in vivo. Absence of GCN2 in C. albicans renders C. elegans resistant to infection-induced lethality (Figure 1A) and absence of GCN4 partially abolish the virulence of C. albicans (Figure 1B).
Interestingly, growth conditions used in our assays induces the filamentation of wild type C. albicans (Figure 2) corroborating the decreased survival rates observed in C. elegans after infection, since C. albicans is converted in a more virulent form in our tested conditions. In addition, resistance of N2 worms to infection with C. albicans GCN2 or GCN4 knockouts is linked to strain filamentation ability. We show that C. albicans GCN4 knockout has decreased filamentous growth (Figure 2E, 2F) and slightly reduced virulence to C. elegans, while GCN2 knockout show slow-growth phenotype and hyphae absence (Figure 2H, 2I) that correlates with marked C. elegans resistance to infection when compared to worms infected with wild type C. albicans (Figure 1A).
Here, we confirmed that Gcn2 loss of function in C. albicans change yeast morphogenesis reducing the biofilm formation capacity, as reported previously [14,29] and renders yeasts less virulent to the invertebrate model of infection C. elegans. Tripathi and co-workers showed that Gcn2 and Gcn4 control C. albicans morphogenesis during amino acid starvation [29] and in this work we show that Gcn2 and Gcn4 loss of function are important to yeast virulence and morphogenesis in the presence of amino acids. In NGM medium there is no glucose supplementation and amino acids provided by bacto-peptone are the only carbon source. Vylkova and co-workers have shown that C. albicans growing in conditions in which amino acids are the primary carbon source result in generation of large quantities of ammonia, raising the extracellular pH and inducing the hyphal switch in vitro [31]. Also, amino acids induce C. albicans phagosomal pH neutralization, hyphal morphogenesis, and escape from macrophages [32]. Considering these data along with our findings, we demonstrate that C. elegans infection with C. albicans in solid NGM could be a valuable tool to study pathogen virulence traits and host responses.
Taken together, we show eIF2-mediated translation regulation modulates C. albicans virulence to C. elegans through Gcn2 function in morphogenesis during amino acid rich conditions. More studies are necessary to address how Gcn2 and Gcn4 regulate and/or integrate with previously described morphogenesis pathways in C. albicans.
Accession numbers
Accession number for C. albicans genes GCN2 (C7_01330C) and GCN4 (C2_09940W) accessed at http://www.candidagenome.org.
Acknowledgements
We thank Caenorhabditis Genetic stock Center (CGC) funded by NIH Office of Research Infrastructure Programs (P40 OD010440) for the N2 Bristol strain. We thank Professor Alistail Brown (Aberdeen University) for C. albicans strains. VSA is funded by the Centro Nacional de Desenvolvimento Cientifico e Tecnologico - CNPq, Brasil (Post-doctoral fellowship - Science without Borders Program - CsF).
Author contributions
VSA and EM conceived and designed the experiments. VSA performed the experiments. VSA and EM analyzed the data and wrote the paper.
Competing interests
The authors have declared that no competing interests exist.
References
- San-Blas G, Travassos LR, Fries BC, Goldman DL, Casadevall A, et al. (2000) Fungal morphogenesis and virulence. Med Mycol 38: 79-86. [Crossref]
- Karkowska-Kuleta J, Rapala-Kozik M, Kozik A (2009) Fungi pathogenic to humans: molecular bases of virulence of Candida albicans, Cryptococcus neoformans and Aspergillus fumigatus. Acta Biochim Pol 56: 211-224. [Crossref]
- Mitchell AP (1998) Dimorphism and virulence in Candida albicans. Curr Opin Microbiol 1: 687-692. [Crossref]
- Rooney PJ, Klein BS (2002) Linking fungal morphogenesis with virulence. Cell Microbiol 4: 127-37. [Crossref]
- Gow NA, van de Veerdonk FL, Brown AJ, Netea MG (2012) Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat Rev Microbiol 10: 112-122. [Crossref]
- Mayer FL, Wilson D, Hube B (2013) Candida albicans pathogenicity mechanisms. Virulence 4: 119-128. [Crossref]
- Biswas S, Van Dijck P, Datta A (2007) Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol Mol Biol Rev 71: 348-376. [Crossref]
- Sudbery P (2007) Mo2021 Copyright OAT. All rights reservn requires septin phosphorylation. Dev Cell 13: 315-316. [Crossref]
- Albuquerque P, Casadevall A (2012) Quorum sensing in fungi--a review. Med Mycol 50: 337-345. [Crossref]
- Ernst JF (2000) Transcription factors in Candida albicans - environmental control of morphogenesis. Microbiology 146: 1763-1774. [Crossref]
- Whiteway M (2000) Transcriptional control of cell type and morphogenesis in Candida albicans. Curr Opin Microbiol 3: 582-588. [Crossref]
- Brown AJ, Barelle CJ, Budge S, Duncan J, Harris S, et al. (2000) Gene regulation during morphogenesis in Candida albicans. Contrib Microbiol 5: 112-125. [Crossref]
- Whiteway M, Bachewich C (2007) Morphogenesis in Candida albicans. Annu Rev Microbiol 61: 529-553. [Crossref]
- Tournu H, Tripathi G, Bertram G, Macaskill S, Mavor A, et al. (2005) Global role of the protein kinase Gcn2 in the human pathogen Candida albicans. Eukaryot Cell 4: 1687-1696. [Crossref]
- García-Sánchez S, Aubert S, Iraqui I, Janbon G, Ghigo JM, et al. (2004) Candida albicans biofilms: a developmental state associated with specific and stable gene expression patterns. Eukaryot Cell 3: 536-545. [Crossref]
- Pukkila-Worley R, Peleg AY, Tampakakis E, Mylonakis E (2009) Candida albicans hyphal formation and virulence assessed using a Caenorhabditis elegans infection model. Eukaryot Cell 8: 1750-1758. [Crossref]
- Pukkila-Worley R, Mylonakis E (2010) From the outside in and the inside out: Antifungal immune responses in Caenorhabditis elegans. Virulence 1: 111-112. [Crossref]
- Pukkila-Worley R, Ausubel FM, Mylonakis E (2011) Candida albicans infection of Caenorhabditis elegans induces antifungal immune defenses. PLoS Pathog 7: e1002074. [Crossref]
- Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77: 71-94. [Crossref]
- Stiernagle T (2006) Maintenance of C. elegans. WormBook 2006: 1-11. [Crossref]
- Pierce CG, Uppuluri P, Tummala S, Lopez-Ribot JL (2010) A 96 well microtiter plate-based method for monitoring formation and antifungal susceptibility testing of Candida albicans biofilms. J Vis Exp 44. pii: 2287. [Crossref]
- Spriggs KA, Bushell M, Willis AE (2010) Translational regulation of gene expression during conditions of cell stress. Mol Cell 40: 228-237. [Crossref]
- Hershey JW, Sonenberg N, Mathews MB (2012) Principles of translational control: an overview. Cold Spring Harb Perspect Biol 4. [Crossref]
- Chow C, Cloutier S, Dumas C, Chou MN, Papadopoulou B (2011) Promastigote to amastigote differentiation of Leishmania is markedly delayed in the absence of PERK eIF2alpha kinase-dependent eIF2alpha phosphorylation. Cell Microbiol 13: 1059-1077. [Crossref]
- Narasimhan J, Joyce BR, Naguleswaran A, Smith AT, Livingston MR, et al. (2008) Translation regulation by eukaryotic initiation factor-2 kinases in the development of latent cysts in Toxoplasma gondii. J Biol Chem 283: 16591-1601. [Crossref]
- Joyce BR, Queener SF, Wek RC, Sullivan WJ, Jr (2010) Phosphorylation of eukaryotic initiation factor-2{alpha} promotes the extracellular survival of obligate intracellular parasite Toxoplasma gondii. Proc Natl Acad Sci U S A 107: 17200-17205. [Crossref]
- Zhang M, Fennell C, Ranford-Cartwright L, Sakthivel R, Gueirard P, et al. (2010) The Plasmodium eukaryotic initiation factor-2alpha kinase IK2 controls the latency of sporozoites in the mosquito salivary glands. J Exp Med 207: 1465-1474. [Crossref]
- Kamthan M, Mukhopadhyay G, Chakraborty N, Chakraborty S, Datta A (2012) Quantitative proteomics and metabolomics approaches to demonstrate N-acetyl-D-glucosamine inducible amino acid deprivation response as morphological switch in Candida albicans. Fungal Genet Biol 49: 369-378. [Crossref]
- Tripathi G, Wiltshire C, Macaskill S, Tournu H, Budge S, et al. (2002) Gcn4 co-ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans. EMBO J 21: 5448-5456. [Crossref]
- Sundaram A, Grant CM (2014) Oxidant-specific regulation of protein synthesis in Candida albicans. Fungal Genet Biol 67: 15-23. [Crossref]
- Vylkova S, Carman AJ, Danhof HA, Collette JR, Zhou H, et al. The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. MBio 2: e00055-11. [Crossref]
- Vylkova S, Lorenz MC. Modulation of phagosomal pH by Candida albicans promotes hyphal morphogenesis and requires Stp2p, a regulator of amino acid transport. PLoS Pathog 10: e1003995. [Crossref]



