Breast Cancer (BC) is the most prevalent cancer affecting females and the leading cause of death around the world. The World Health Organization (WHO) has announced that the incidence of BC increases approximately 1.8-2 % annually. Hence, it is important that new prevention and treatment strategies involving epigenetics and nutrigenomics are explored. Epigenetics refers to potential changes in gene expression and chromatin structure without alteration of DNA sequence, but still modulates the expression of a particular phenotype. Nutrigenomics determines the effect of dietary habits on cancer risk and tumor behavior, both in progression and inhibition of cancer. The modulation of chromatin structure is an essential component in the regulation of transcriptional activation and repression. Therefore, identifying the regulators of gene expression changes during cancer progression is essential for early diagnosis and inhibition of this malignancy. The methylation of promoter genes, as well as the interplay between microRNAs (miRNAs) and messenger RNAs (mRNAs) of target genes, is the primary component of epigenetics. Any defect in these processes is considered a crucial mechanism for gene and pathway dysregulation in all human cancers, including BC. Nutritional genomic and epigenetic mechanisms may play a pivotal role in prevention as well as early diagnosis of BC, especially for closely related female family members of BC patients. It seems cytosine methylation in Cytosine-phosphate-Guanine dinucleotide (CpG) Islands reflects changes in balance tissues rigidity. Hypo- and hyper-methylation of CpG Islands (CGIs) play a crucial role in development of BC via up-regulation of oncogenes and down-regulation of tumor suppressor genes (TSGs). These could be the most effective mechanisms to distinguish BC from other types of cancer.
cancer, breast cancer, nutrigenomics, DNA methylation, epigenetics, miRNA
In 2018, with a 24.2% incidence rate and 15% mortality rate, breast cancer (BC) was the most commonly diagnosed cancer in women [1]. Epidemiological surveys have elucidated that individual risk increases with the number of first and second-degree female relatives diagnosed with BC, as well as the age of their disorder onset, compared with the general population [2,3].
Relatives of BC patients are 2 to 4 times more likely at risk than general population concerning BC [4,5]. The most understood forms of genetic alteration involve either mutation, amplification or deletion within the genome. Traditionally, it has become clear that the molecular mechanisms underlying the development and progression of cancers, including BC, involve gene modification and regulation beyond changes to the DNA sequence. More recently, it was brought forth that epigenetic mechanisms, in which no changes to the DNA sequence occurs but instead gene expression patterns are altered, dysregulate information inside the genome of a healthy cell, leading to malignancy such as cancer [6]. Moreover, hypo- and hyper-methylation of CGIs play a crucial role in development of BC via up-regulation of oncogenes and down-regulation of TSGs, respectively [7]. Even more recently, nutritional genomics is the latest method of investigation of gene modulation, and it appears that diet and the gut can cause significant malignancy [8,9]. Nutrigenomics can be used to understand the effect of nutrients on DNA/gene expression and the proteome and metabolome, as well as the effects of diet-gene interplay in the preservation of health conditions or development of diseases such as cancer [10,11].
It has been elucidated that many essential and non-essential nutrients such as micronutrients
(e.g., vitamins and minerals), macronutrients (e.g., fatty acids and proteins) and some bio-reactive chemicals (e.g., flavonoids, carotenoids, phytosterols, eicosapentaenoic acid, and docosahexaenoic acid) can adjust gene expression during physiological and pathological processes via epigenetic mechanisms [12,13]. Nutritional genomics may also provide new information to develop treatments of diseases at the cellular level and preventative measures through the diet. Although it is challenging to determine the exact role of nutrients in epigenetic mechanisms because diet is often accompanied with dependent lifestyle factors such as poor diet, inappropriate body mass index (BMI), insufficient or lack of physical activity, and the use of drugs, alcohol or tobacco, there is accumulating evidence on the positive effects of food habits on reduction of cancer risk [14,15], and researchers have indicated that dietary and behavioral factors have a significant role in approximately one-third of cancer-related deaths [16,17].
In the current study, we 1) discuss the role of DNA methylation and miRNAs in the occurrence of BC in relation to oncogene activation and tumor suppressor gene (TSG) inactivation, and 2) highlight the beneficial effects of some nutritional components on the function and structure of non-coding RNA and DNA methylation. We also discuss the role of nutrigenomics as a non-invasive method to explore the epigenetic mechanisms involved in BC and also in the prevention, treatment, early diagnosis, and distinguishing a variety of cancers from each other. In addition, the importance of non-coding RNAs, including miRNAs in body fluids, also need to be further clarified.
DNA methylation is an epigenetic mechanism which almost modulating the function of the genes and impacting on gene expression, thereby it has a significant role in cellular differentiation, diversity of gene expression, as well as the development of nearly all types of cancer [18,19]. DNA methylation is a post-replicative process in which a methyl group is added at the 5-carbon of the cytosine ring, characterized as 5-methylcytosine (5-mC). These methyl groups project into the major groove of DNA and inhibit transcription. Most cytosine methylation occurs in the sequence context 5'CG3' and happens almost exclusively at cytosines that are followed immediately by a Guanine (CpG, Cytosine-phosphate-Guanine dinucleotide) [5]. The methyl transferase family catalyzes the methylation process, when a methyl group is transmitted from S-adenosyl methionine (SAM) to the cytosine. DNA methyl transferase 1 (DNMT1) is responsible for the maintenance of established patterns of DNA methylation, and DNMT3a and DNMT3b mediate establishment of de novo DNA methylation patterns (Figure 1) [20].
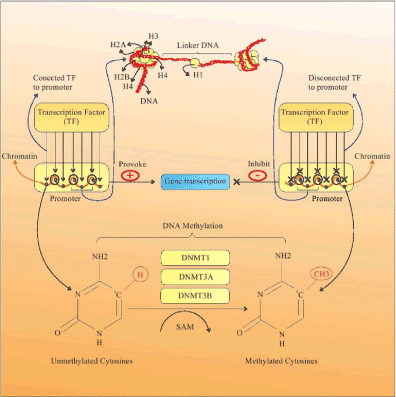
Figure 1. DNA methylation. DNA methylation is a post replicative process that results from transfer of a methyl (CH3) group to the 5-carbon of the cytosine (C5) ring by DNMTs, and is characterized as 5 methylcytosine (5-MC). Hyper-methylation of CGIs in gene promoters leads to alterations in chromatin structure and restricts access of TFs to the gene promoter, quelling the gene transcriptions and silencing of the gene. DNMT: DNA methyl transferase; TF: Transcription factor; CpG Islands
DNA methylation can occur in four types of regions including: 1) repetitive sequences, 2) CGI Promoters, 3) CGI Shore, and 4) the gene body (coding sequences) [21]. CGIs are located in approximately 60 percent of the repetitive DNA sequences and human gene promoters [22]. In constant regions, the CpG dinucleotides mostly tend to stay methylated, whilst in gene promoters, the CpGs are mostly unmethylated to prevent chromosome instability. In the first three cases, DNA methylation is typically related to quell of gene expression and transcription. However, DNA methylation can occur in the gene body rather than the promoters [21,23].
The expression of genes is facilitated by engagement of transcription factors (TF) to the gene promoter DNA [24]. However, the process of methylation leads to repression through changes in chromatin structure restricting access of TF to the gene promoter, preventing gene transcription and expression. Hyper-methylation of DNA causes the gene to be silenced (Figure 1) [20].
DNA methylation patterns vary profoundly among healthy and cancerous cells (Figure 2). For instance the total 5-methylcytosine can be 20-60% less in repetitive elements of cancerous cells compared to healthy cells, resulting in chromosomal instability [21,25]. Hypo-methylation also occurs in specific regional DNA (promoters) of cancerous cells, resulting in the activation of potential oncogenes, as well as stimulating loss of imprinting at other DNA locations [25,26]. Conversely, hyper-methylation of CGI promoters of genes central to DNA repair and cell cycle control pathways correlates with very serious tumor disease in humans. Moreover, tumor cells express atypical DNA methylation at CGI shores, and sometimes a specific enzyme that triggers the detachment of the methyl group from methylated DNA [27].
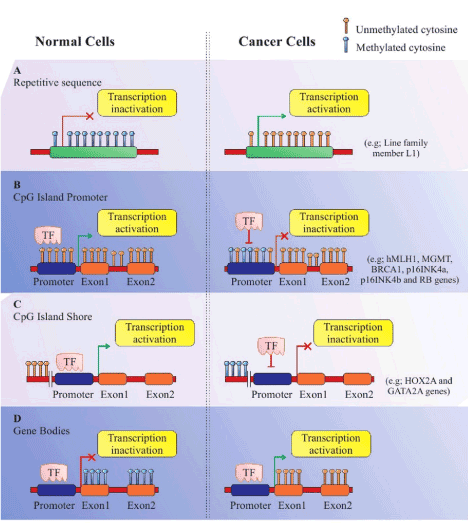
Figure 2. DNA methylation templates in normal and cancerous cells. (A) Repetitive Sequence: Hyper-methylation in repetitive sequences prohibit gene transcription in healthy cells, while reduction of 20 to 60 percent in the 5-methylcytosine in cancerous cells leads to deformation of chromatins and triggers gene transcription. (B) CpG Island Promoters: CpG island promoters are generally hypo-methylated and activate transcription in healthy cells. Conversely, CpG island promoters are hyper-methylated in cancerous cells, resulting in transcriptional inactivation. (C) CpG Island Shore: The mechanism of DNA methylation templates is the same as in B, except that the CpG island shore is located up-stream. (D) Gene Bodies: CpG methylation in gene bodies prohibit gene expression in healthy cells, but is reversed in cancer cells, resulting in initiation of transcription at several incorrect loci
BC is a heterogeneous disease and has two main histological subtypes: ductal and lobular adenocarcinomas. These subtypes are distinct due to the clinical presentation and behavior, and morphological and molecular characteristics [6,28]. In recent years, DNA methylation patterns of target genes have been considered by investigators as a biomarker for BC prognosis (Table 1). Kim et al. examined the status of three genes; LHX2, WT1 and OTP, in primary tumors and adjacent and other healthy tissues, and found that the frequency of aberrant hyper-methylation in primary tumors (LHX2= 43.6%, WT1= 89.7% and OTP= 100%) was high. However, the methylation frequencies were intermediate and rare in adjacent healthy tissue and other healthy tissue [29]. DeRoo et al. assessed Line-1 methylation in blood samples, and found a dose dependent and negative correlation between under-methylation of Line-1 and risk of BC in non-hispanic caucasian women. They also found that three Single Nucleotide Polymorphism (SNP) genes; SLC19A1 (rs1051266), MTRR (rs10380) and MTHFR (rs1537514), related to Line-1 methylation, and MTHFR (rs1537514) is also linked to BC [30]. Chimonidou et al. indicated that in Cell-Free DNA (CF-DNA) of BC patients, CST6 promoters were hyper-methylated compared to healthy individuals. For this reason, they suggested that CST6-promoter methylation, especially in CF-DNA of BC patients, could potentially be used as a novel plasma tumor biomarker for BC [31]. Rauscher et al. studied DNA regions including promoters (BRCA1, CD44, ESR1, GSTM2, GSTP1, MAGEA1, MSI1, NFE2L3, RASSF1A, RUNX3, SIX3 and TFF1), far upstream regions (EN1, PAX3, PITX2, and SGK1), introns (APC, EGFR, LHX2, RFX1 and SOX9) and repetitive sequences (Line-1 and satellite 2), and found that most of these gene regions were hyper-methylated in BC compared to adjacent tissue, except TFF1 and MAGEA1 gene regions which were hypo-methylated. They also investigated six gene regions (RASSF1, TFF1, EGFR, SGK1, LINE-1 and satellite 2) using a large sample size (105-129 patients & 15-18 reduction mammoplasty), and indicated aberrant methylation in adjacent, histologically healthy tissues compared to non-cancerous mammoplasty samples [32]. Stefansson et al. assessed two biologically distinct and aggressive BC subtypes: Basal-like and Luminal-B. DNA methylation patterns in Basal-like and Luminal-B were distinguished by hypo-methylation of gene bodies and methylation of CGI promoters [33]. Tian et al. have shown that FOXF2 levels are augmented in a Basal-like BC subtype, and acts as an epithelial-mesenchymal transition suppressor. They explained that in BC cells, FOXF2 has a novel function as a transcriptional target of SP1. Through methylation of CGI, FOXF2 proximal promoter region abolished SP1 binding, resulting to the suppressing of FOXF2 expression. It seems that, simultaneously with DNA, methylation and abrogation of SP1 binding to the proximal promoter region, suppression of progression and promotion of proliferation as a dual function of FOXF2 in related to SP1 will be influenced; therefore, the expression of FOXF2 will be silenced in BC cells [34]. Another study assessed DNA methylation profiles in breast tumors of women with a strong BC family history compared to adjacent non-tumor tissues, and selected 40 hyper-methylated genes from The Cancer Genome Atlas (TCGA) based on breast tumor samples [35]. They demonstrated that only 32.5 percent of these genes including SEPW1, PCDHGA4, CCDC36, C1orf14, RPTOR, C1orf114, ZNF454, USP44, CSMD3, PRKAR1B, SLC7A14, SOX2OT and RYR2, were highly methylated in their breast tumor samples compared to the adjacent histologically healthy tissues. Accordingly, they suggested that identifying methylation markers as a biomarker for early diagnosis of BC cases in particularly high risk groups, like familial cases, can be examined [35]. Dehbid et al. suggested that the hyper-methylation of MAP9 gene in BC patients causes the reduced gene expression consistently seen in these patients. Reduced gene expression of MAP9 in BC leads to instability of P53, which can inhibit apoptosis of the cancer cells. Therefore, Hyper-methylation of MAP9 gene and subsequent destabilization of P53 may be useful molecular biomarkers for BC diagnosis [36].
Table 1. Summary of miRNAs, DNA methylation and BC studies
References |
Samples |
Methods |
Major finding |
Summary of DNA methylation studies |
Kim et al., 2012 [29] |
- BC cell lines: MCF-7 (HTB-22) and MDA-MB-231 (HTB-26).
-Fresh-frozen tissue specimens.
- Three candidate genes for methylation status in primary tumor, normal tissues, and paired adjacent normal appearing tissues (n=39):
LHX2, WT1 and OTP. |
- CpG microarray analysis: MeDIA + MBD2bt
- The hybridized images: analyzed by Agilent DNA microarray Scanner
- Bisulfite PCR and pyrosequencing primers: augment 3 to 5 CpG dinucleotide sites in the target areas of genes. |
- Aberrant hyper-methylation of genes in primary tumors (43.6% for LHX2, 89.7% for WT1 and 100% for OTP), whereas frequencies were invisible in normal tissues and intermediate in paired adjacent normal tissues.
- OTP gene most probably have a predominately role in breast carcinogenesis. |
DeRoo et al., 2014 [30] |
- A prospective cohort study among 50 884 (294 cases and 646 non-cases; ages: 35-75 years).
- LINE-1 methylation
was assessed in DNA from peripheral blood samples and blood clots. |
- Global methylation was gauged by pyrosequencing of the LINE-1 repetitive element. |
- Three SNP in genes are related to LINE-1 methylation: MTRR (rs10380), SLC19A1 (rs1051266), and MTHFR (rs1537514).
- MTHFR (rs1537514) was connected with BC risk.
- PON1 (rs757158) was associated with BC but not methylation. |
Chimonidou et al., 2013 [31] |
- 233 clinical samples:
a) One pilot testing group: 27 patients, 46 patients for verified metastasis & 37 healthy female (blood donors).
b) Independent cohort: 123 patients.
- The BC cell line: MCF 7 (positive control in MSP). |
-The methylation status of CST6 in circulating cfDNA was assessed by nested MSP. |
- CST6 promoter is crucially methylated in cfDNA of BC participants compared to healthy individuals.
- CST6 promoter methylation in cfDNA: recognized as a newfound plasma tumor biomarker for BC. |
Rauscher et al., 2015 [32] |
- BC patients: n = 129 (ages: 30-79).
- Control: 18 unaffected, cancer-free patients.
- The DNA regions studied were promoters, far-upstream regions, introns, and the LINE-1 and satellite 2 DNA repeats. |
- Pyrosequencing was conducted on bisulfite-treated DNA from FFPE |
- The majority of the assessed gene regions: hyper-methylated in cancer vs. adjacent tissue, except for TFF1 and MAGEA1 regions were highly hypo-methylated.
- Confirming frequent DNA methylation changes in invasive BC at a various genome location. |
Stefansson et al., 2015 [33] |
- Samples: 25 normal breast tissue (adjacent to tumor lesions of patients) and 350 primary invasive breast tumors. |
- DNA isolation: phenolchloroform/Proteinase-K method.
- DNA methylation analysis:Infinium HumanMethylation450 BeadChips.
- The PyroMark Q96 system for pyrosequencing was applied to
evaluate. |
-Discovered signatures related to two biologically distinct BC subtypes: Basal-like and Luminal-B.
-DNA methylation patterns linked to the Luminal-B subtype: CpG island promoter methylation.
-Basal-like tumors: hypo-methylation of gene body. |
Tian et al., 2015 [34] |
-BC cell lines: MCF-7, MDAMB-453, and MDA-MB-231 and the human mammary epithelial cell line: MCF-10A.
- 20 primary BC tissue specimens (invasive ductal carcinoma).
- DNA achieved from cultured cells or frozen tissue. |
-Using SssI, HhaI, and HpaII methylases or no enzyme (mock): Influence of CpG Island Methylation Status on FOXF2.
- Cell proliferation: 3-(4,5-dimethyl- thiazol-2-yl)- 2,5-diphenyl- tetrazolium bromide (MTT) assay
- Invasion and migration of breast cells: Matrigel-coated and uncoated Transwell, respectively. |
- CpG island methylation of the FOXF2 proximal promoter: engaged in the regulatory process of the subtype-specific expression of FOXF2 in BC cells.
-SP1: adjusted the transcriptional activity of FOXF2 by straight binding to the
proximal promoter region.
- Function and expression of FOXF2 in BC cells modulated through the composed impacts of DNA methylation and SP1 transcriptional regulation. |
Wu et al., 2017 [35] |
- 40 FFPE breast tumors and adjacent non-tumor tissues from BC cases (have family history of BC).
- The genes had selected from analysis of TCGA project. |
- DNA methylation: Illumina MiSEQ instrument (measuring DNA methylation in candidate genes.) |
-13 out of 40 genes were highly methylated.
- In regard to TCGA, methylation levels in SEPW1, PCDHGA4, CCDC36, C1orf14, RPTOR, C1orf114, ZNF454, USP44, CSMD3, PRKAR1B, SLC7A14, SOX2OT and RYR2 were remarkably observed in the breast tumor.
- 32% of tested genes: hyper-methylated in BCFR (36.1%: SEPW1, 10%: RYR2). |
Dehbid et al., 2016 [36] |
Samples: 30 healthy women and 30 BC women. |
- DNA isolated from 200 µl whole blood by MBST DNA extraction Kit.
- Detecting DNA with CPG island methylation by restriction endonuclease quantitative PCR method. |
- Hyper-methylation of MAP9 gene in BC can be used as molecular biomarker for BC diagnosis. |
Summary of miRNAs studies |
Ng et al., 2013 [39] |
- BC: n= 260 (clinically diagnosed),
- Normal subjects: n=170 (pathologically verified not to have BC and no history of other cancers).
- MiRNA profiling was proceeded in four groups: 1) BC plasma, 2) normal plasma, 3) tumor, and 4) adjacent non-tumor tissue. |
- Extraction total RNA from tissues: TRIzol reagent
- Extraction total RNA containing small RNA from 500 ml of plasma: Trizol LS reagent and miRNeasy Mini Kit.
-Plasma miRNA Profiling: using TaqMan Array Human miRNA.
- Plasma miRNA Quantitative PCR: SYBR green qRT-PCR assay |
- Crucial promotion of the miRNA- 16, miRNA-21, and miRNA-451 and remarkable decline of miRNA-145 in the plasma of BC patients.
- The composition of plasma miRNA-145 and miRNA-451 surfaces indicated the best markers for BC prediction. |
Cuk et al.,2013 [40] |
- Blood samples were prepared from 80 healthy and 127 female sporadic BC.
- Malignant tissue samples were provided from non-neoadjuvant BC patients during surgery. |
- Extraction total RNA from 400 ml of plasma: Trizol LS reagent and miRNeasy Mini Kit.
-Plasma miRNA Profiling: TLDA.
-Validation of chosen marker candidates: RT reactions were done by using TaqMan MicroRNA RT Kit and miRNA specific RT primers. |
- Four miRNAs (miRNA-148b, miRNA-376c, miRNA-409-3p and miRNA-801) significantly up-regulated in the plasma of BC patients.
- Based on ROC curve, only three miRNAs (miRNA-148b, miRNA-409-3p and miRNA-801) had an equal discriminatory power among BC samples and healthy controls. |
Si et al., 2013 [41] |
- Tumor tissues, paired NATs, and matching serum samples were collected from 48 newly diagnosed breast carcinomas.
- Other serum samples: 52 cases of BC and 20 of HC. |
- Total RNA isolation: The miRNeasy Mini Kit and mirVana PARIS Kit.
- MiRNA quantification: SYBR green qRT-PCR assay. |
- MiRNA-92a and miRNA-21 expression may be used to discriminate the BC patients from the HC.
- Circulating miRNA-92a has potential use as novel BC biomarker.
- Decreased levels of miRNA-92a and increased levels of miRNA-21 were connected with tumor size and a positive lymph node status in compared to clinicopathologic data of the BC patients. |
Chan et al., 2013 [42] |
- Extracted miRNA from paired fresh frozen BC tumors, normal tissue, and serum samples of patients (n=32).
- Serum samples from normal controls (n = 22).
- Additional serum samples from patients with BC (n = 132) and control serum samples (n = 101). |
- MiRNA extraction: From tissue or serum samples by miRVana or miRNeasy kits, respectively.
- Quality control of RNA from tissue samples: Agilent Bioanalyzer.
-Quality control of serum samples: singleplex LNA RT-PCR. |
- Only 7 miRNAs were upregulated in both serum and tumors.
- MiRNA-1, miRNA 92a, miRNA-133a, and miRNA-133b identified as the most significant diagnostic markers.
- Serum-based miRNA signatures related to BC were successfully validated and derived. |
Sochor et al., 2014 [43] |
- EBC Patients sera = 63 and HC= 21.
- Timing of sera collection: one-day prior operation (time point I), 14–28 days after operation before any non-surgical treatment (time point II) and 14–28 days after first treatment modality (time point III). Upon clinical relapse the patients (N = 3) donated another serum sample (time point IV). |
- RNA extraction: miRNeasy® Mini Kit.
- RT was done by using High Capacity cDNA Reverse Transcription Kit supplemented with miRNA-specific primers. |
- Surgery had drastically effect on three serum oncomiR (miRN A-155, miRNA-181b, and miRNA-24) levels, whilst adjuvant therapy mainly reduced all of four oncomiR (miRNA-155, miRNA-181b, miRNA-24 & miRNA-19a). |
Erbes et al., 2015 [44] |
- MSU gathered in a case–control cohort of:
1) 24 untreated patients, 2) newly diagnosed with primary BC, 3) 24 HCs.
- Serum samples of four consecutive patients and HCs. |
- RNA isolation:
Urine: Norgen’s Urine microRNA Purification Kit, Serum: Norgen’s kit.
- Pre-amplification: augment of miRNA-cDNA quantity was attained by application of Megaplex™ PreAmp Primers, Human Pool A.
- MiRNA expression levels: quantitative real-time-PCR applying TaqMan® MicroRNA Assays. |
- MiRNA-21, miRNA-125b, miRNA-451 and miRNA-155altered and regulated in BC patients in comparison with HCs.
- MiRNA 155 levels were higher, while miR-21, miRNA-125b and miR-451 levels were low in BC patients compared to HC.
- May be urinary miRNAs play a potential role as non-invasive innovative urine-based biomarkers for BC detection. |
Shimomura et al., 2016 [45] |
- Sera samples:
BC= 1280, NCC= 2836, other types of cancer= 451, non-breast benign diseases= 63.
- The samples divided into: 1) Training cohort (NCCs, other cancers and BC)
2) Test cohort (NCCs & BC). |
- Total RNA: extracted from a 300 µL serum sample using 3D Gene RNA extraction reagent from a liquid sample kit.
- Comprehensive miRNA expression analysis: done by using a 3D-Gene miRNA Labeling kit and a 3D-Gene Human miRNA Oligo Chip.
- miRNA to be present:
The corresponding microarray signal was more than the (mean + 2 × standard deviation) signal of the negative controls.
- Quantitative RT-PCR assay: TaqMan microRNA assay |
- A collection of miRNA-1246, miRNA-1307-3p, miRNA-4634, miRNA-6861-5p, and miRNA-6875-5p assessed from serum can be used to survey EBC and to differentiate BC from pancreas ⁄ biliary tract ⁄ prostate benign diseases or other cancers. |
Tsai et al., 2018 [46] |
- First stage:
10 samples from 7 young BC patients (tumor: n = 6, normal tissue: n = 4) have been analyzed by an Agilent microRNA 470 probe microarray.
- Second stage:
85 candidate miRNAs have been validated in 145 BC in 4 subgroups:
1- Very young BC (n=15, ≤35 y)
2- Young BC (n=37, 36-40 y)
3- Premenopausal BC (n= 47, 41-50 y)
4- Postmenopausal BC (n=46, >50 y) |
- Total RNAs have been extracted from tissue by using TRIzol.
- 85 miRNAs were selected for qPCR by Agilent microRNA 470 probe microarray through two ways:
1- MiRNAs that the fold-change and the t-test p value were > 1.5 and < 0.05 between tumor and normal breast tissues (44 miRNAs).
2- MiRNAs that have been reported
in the literature to be related to BC (41 miRNAs). |
- 6 miRNAs had significantly differential expression, 2 out of 6 have been down-regulated, including miRNA-145 and miRNA-335, whilst 4 out of 6 have been up-regulated which consist of miRNA-21, miRNA-200a, miRNA-200c and miRNA-141.
- As well as, the below detections have been seen separately in each subgroup:
1- In very young BC 3 miRNAs (miRNA-96, miRNA-182, and miRNA-183) and 8 miRNAs (miRNA-10a, miRNA-10b, miRNA-125b, miRNA-127-3p, miRNA-130a, miRNA-143, miRNA-19, and miRNA-320) have been down- and up-regulated, respectively.
2- For young BC, upregulation of 3 miRNAs (miRNA-30b, miRNA-30d, and miRNA-149) and downregulation of 3 miRNAs (miRNA-487b, miRNA-548c-5p, and miRNA-181d) have been observed.
3- Only miRNA-206 has been down-regulated in premenopausal BC.
4- No alteration has been observed in postmenopausal BC. |
MeDIA: Methylated DNA isolation assay; MBD2bt: Methyl DNA binding domain; MeDIP: Methyl cytosine DNA Immunoprecipitation; BC: Breast cancer; CGI: CPG island; ERα-: Estrogen receptor α-negative; MSP: Methylation-specific PCR; cfDNA: Cell-free DNA; HER2: Tyrosine kinase-type cell surface receptor; TNBC: Triple-negative breast cancer; HRD: Homologous recombination deficiency; FFPE: Formalin-fixed, paraffin-embedded sections; FOXF2: Fork-head box F2; BCFR: Breast cancer family registry; SNP: Single-nucleotide polymorphisms; TCGA: The cancer genome atlas; ROC: Receiver operating characteristic; TLDA: TaqMan low-density arrays; FDR: False discovery rate; RT: Reverse transcription; NATs: Normal adjacent tissues; HC: Healthy control; LNA RT-PCR: Locked nucleic acid real-time PCR; EBC: Early-stage breast cancer; MSU: Midstream specimen of urine; NCC: Non-cancer control; qPCR: Quantitative real-time PCR analysis
There are several atypical methylation patterns and subsequent changes in gene expression linked to BC that have proven useful for early detection, risk assessment and characterization of BC this last decade. Since DNA methylation is more stable and long-lasting than previously thought, and known to regulate gene expression triggering cells to become cancerous, it is now considered possible to use methylation patterns of genes involved in BC predisposition and development as biomarkers to diagnose patients most susceptible such as first relatives of patients [37-46].
The process by which miRNAs adjust gene expression is complex, since one miRNA can target thousand numbers of mRNAs, and one mRNA can be modulated by various miRNAs. MiRNAs are small (18-24 nucleotides long) non-coding RNAs that regulate gene expression at the post-transcriptional level by attaching to the target (3´-UTR) of mRNA. Production of miRNA is completed in the nucleus and cytoplasm of cells [47, 48]. In the nucleus, transcription of primary miRNAs (pri-miRNAs), 1-3 Kb long, are produced by RNA polymerase II from individual genes containing their promoter or intragenically from protein-coding genes [49]. The pri-miRNAs are then cleaved into stem-loop precursor miRNAs (pre-miRNA), 70-100 nucleotides long, by Drosha (RNA polymerase III) and DGCR8 cofactor (double-stranded RNA binding protein). Pre-miRNAs are transported to the cytoplasm by binding to Exportin-5 receptors (exporting-5-RanGTP), and then processed into a mature miRNA duplex by Dicer (RNase III enzyme), Trans Activating Response RNA-Binding Protein 2 (TARBP2) and AGO2 (DICER complex) [50]. The duplex strands are then separated and the complementary strand can be loaded in the RNA-induced silencing complex (RISC) or can also be degraded [51]. Recent findings show that the duplex strands have considerable biological functions, whereas the mature miRNAs (the guide strand) is incorporated into the RISC, including the GW182 and Argonaut (AGO) proteins. A special 6-8 nucleotide sequence at the 5´ end of the miRNA, called seed sequence, determines the role of miRNA in gene regulation. Thereby, the degree of homology between the seed sequences of miRNA and mRNA, triggered at 3´-UTR of the mRNA, delineates mRNA degradation, translational inhibition, or transcriptional activation. Seed matches can occur in any region of mRNA but the highest affinity occurs in the 3´ untranslated region (3´-UTR) of an mRNA (Figure 3) [51-54].
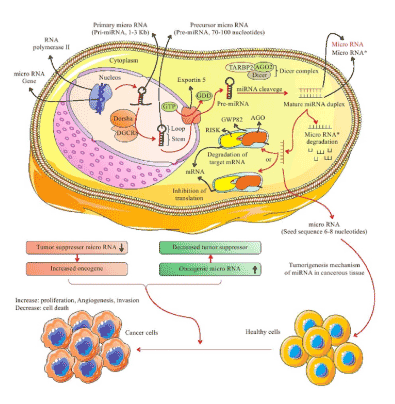
Figure 3. The biogenesis of miRNA. Initially, primary transcription of miRNAs (pri-miRNAs), 1-3 Kb long, are produced by RNA polymerase II. Next, the pri-miRNAs are cleaved into stem-loop pre-miRNA (70-100 nucleotides long) by Drosha (RNA polymerase III) and double-stranded RNA binding protein DGCR8. Pre-miRNAs are transported to the cytoplasm by binding to Exportin-5 receptors (exporting-5-RanGTP) and then processed into a mature miRNA duplex by Dicer (RNase III enzyme), Trans Activating Response RNA-Binding Protein 2 (TARBP2) and AGO2 (DICER complex). The duplex strands are then separated: the complementary strand can be loaded in the RISC or is degraded, and the mature miRNA (the guide strand) is incorporated into the RNA-induced silencing complex (RISC) which includes the GW182 and Argonaut (AGO) proteins. The degree of homology of the miRNA “seed” to the 3′ UTR target sequence of the mRNA determines the mRNA degradation, translational inhibition, or transcriptional activation. Nonetheless, dysregulation of miRNAs expression is able to have a major role in the development of tumor progression, including tumor growth, invasion, proliferation, cell death and angiogenesis. An oncogenic miRNA is intended to repress the translation of a tumor suppressor gene, provoking tumorigenesis, angiogenesis, invasion and leading to tumor formation. Conversely, a tumor suppressor miRNA can prohibit the expression of oncogene. Pri-miRNA: primary miRNA; pre-miRNA: precursor miRNA
There is evidence of an intercellular communication function of miRNAs [55], and some miRNAs have been reported to intervene in gene cell-cycle regulatory processes and effect other proteins involved in cell proliferation [56,57]. Moreover, some miRNAs have a significant effect on tumor suppressor genes, decreasing proliferation and inducing apoptosis, and the levels of these miRNAs are low in cancer cells. In fact, in cancer cells the dysregulation of miRNA expression plays a significant role in the development of tumor progression, including invasion, tumor growth, angiogenesis, and metastatic capability (Figure 3) [58]. In addition, miRNAs are involved in tumor initiation by the regulation of cancer stem cell (CSC) properties, such as tumorigenicity, self-renewal ability, and resistance to different therapies and drugs. Overall, dysregulated miRNAs can act as oncogenic miRNAs (oncomiRs) or TSGs. Moreover, both effects of miRNAs can be identified (Table 2). Chromosomal regions engaged in encoding oncogenic miRNAs may be amplified, resulting in up-regulation of oncogenic miRNAs and simultaneous silencing of TSGs. Alternatively, tumor-suppressive miRNAs are mostly placed in fragile sites along the chromosome, and omission or mutation of these regions leads to a reduction or loss of expression of tumor-suppressive miRNAs and subsequent up-regulation of their target oncogenes [59-77].
Table 2. miRNA and their role in various cancer
|
|
miRNA type |
Cancer type |
Comment |
OncomiRs |
miR17-92 [63]
(miR-17, miR-18a, miR-19a, miR-19b, miR-20a, and miR-92a) |
B-cell lymphoma, colon and gastric cancer. |
Acts as multi-targeted regulators of T-ALL tumor suppressors including PTEN, BIM, NF1, FBXW7, IKZF1 and PHF6 |
miR-21 [64] |
Breast, colon, lung cancer, and glioblastoma, NSCLC. |
Increases growth, promote migration & invasion, and enhance chemo- or radio- resistance, metastasis. |
miR-106 [65] |
Gastric and colorectal cancer |
Proliferation of tumor cells. |
miR-10b [66] |
Breast and Esophageal Cancer |
Cell motility and invasiveness. |
miR-191 [67] |
Human colorectal and BC |
Promoting cell proliferation and migration (by inducing TGF-β2 expression). |
Tumor suppressor miRNAs |
let-7 [68] |
Lung cancer and Burkitt lymphoma |
Engaged in the proliferation, apoptosis and invasion of cancer cells; induced by EphrinA1 and inhibits RAS. |
miR-15a, miR16-1 [69] |
CLL, prostate cancer, and mesothelioma |
MiR-15a: Inhibits growth of MPM cells.
MiR16-1: Downregulates cell cycle and anti-apoptotic genes CCND1 and BCL2. |
miR-29 [70] |
Lung and breast cancer |
MiR-29a reduces the invasive phenotype of cancer (miR-29a expression in the invasive variant was less than 45% compared to non-invasive cell line). |
miR-34a [71] |
Prostate cancer, HCC, and mesothelioma. |
Engaged in the regulation of different cancer processes, such as apoptosis, proliferation, invasion, migration, and metastasis. |
miR-126 [72] |
Lung and breast cancer |
Perhaps acts as a tumor suppressor via prohibition of cell invasion by downregulating ADAM9 (metalloprotease 9). |
Both OncomiRs and Tumor suppressor miRNAs |
miR-24 [73, 74] |
NSCLC (O)
LSCC (TS) |
Acts as oncomir through regulation of its functional target NAIF1.
Expression of miR-24 in Hep2 cells triggered cell morphology alterations and prohibited cell invasion and proliferation ability by targeting S100A8 at the translational level. So through under-expression of S100A8 in LSCC, miR-24 act as TS. |
miR-125 [75]
(hsa-miR-125a, hsa-miR-125b-1 & hsa-miR-125-2) |
Pancreatic, prostate and oligodendro glial cancer (O)
Melanoma, osteosarcoma, ovarian, bladder, and breast cancer (TS) |
Members of the family have main roles in cell proliferation, differentiation, and apoptosis by targeting matrix-metalloprotease, transcription factors, and growth factors. |
miR-155 [76] |
Lymphoma and BC (O)
Melanoma, ovarian and gastric cancer (TS) |
Up-regulation of miR-155 can restrain melanoma growth through down regulation of SKI protein, so improve proliferation and apoptosis.
Also, upregulation of miRNA 155 in SGC-7901 and MKN-45 gastric cancer cells suppressed cell adhesion, invasion, and migration. |
miR-221/222 [77] |
Glioblastoma, HCC, BC, and NSCLC (O)
OTSCC and Erythrocytes (TS) |
MiR-221/222 acts as oncomir through regulation of its functional target P57 and DDIT4 (HCC), Puma (glioblastoma), ER-α and FOXO3 (Breast), PTEN (NSCLC).
MiR-221/222 acts as TS through regulation of its functional target SODI MMP1 (OTSCC), c-Kit (Erythrocytes). |
SCLC: Small cell lung cancer; T-ALL: T-cell acute lymphoblastic leukemia; NSCLC: Non-small cell lung cancer; CLL: Chronic lymphocytic leukemia; MPM: Malignant pleural mesothelioma; HCC: Hepatocellular carcinoma; EMT: Epithelial to mesenchymal transition; LSCC: Laryngeal squamous cell carcinoma; NAIF1: Nuclear apoptosis-inducing factor 1; OTSCC: Tongue squamous cell carcinoma; O: OncomiR; TS: Tumor suppressor miRNA.
There are several studies linking miRNA suppression to BC development, from cancer-initiation and BC metastasis, cell proliferation, angiogenesis, and invasion. Specifically, the expression of Drosha and Dicer enzymes, key to miRNA synthesis, is dysregulated, as is the DGCR8 protein complex [78]. The recently discovered absence of miRNAs in BC cells not only allows diagnosis, prognosis and determination of suitable therapeutic approaches, but may also provide novel targets for therapeutic replacement therapies in cancer diseases, in particular in BC [79]. Following synthesis, stable miRNAs are transferred outside the cell and are readily detectable in peripheral blood samples. These features can play a pivotal role in monitoring secreted miRNA levels from the expanding early-stage BC tissue, and could be a useful novel biomarker for early BC detection [43,44,80].
Ng et al. proposed that circulating miRNAs can be used as specific biomarkers for BC diagnosis. In order to prove this theory, they assessed miRNA profiles in tumor tissue, adjacent non-tumor tissue, corresponding plasma from the same BC patients, and plasma from matched healthy controls. In addition, they validated particular items by a case-control cohort and then blindly verified in an independent set of BC patients and healthy controls. Profiling of miRNAs showed that only three of them, including miRNA-16, miRNA-21 and miRNA-451, significantly increased in both plasma and tissue of BC patients and declined after surgery. Furthermore, miRNA-145 levels significantly decreased in BC patients. Interestingly, they found that the combination of plasma miRNA-451 and miRNA-145 levels was the best biomarker for BC prediction and prevention [39]. A study by Cuk et al. also emphasized the potential role of circulating miRNAs in plasma as early detection markers for BC. They found that four miRNAs; miRNA-148b, miRNA-376c, miRNA-409-3p and miRNA-801, were significantly up-regulated in the plasma of BC patients compared to healthy controls. The authors concluded that the identified miRNAs can be applied as multimarker blood-based tests for early detection of BC [40]. Si et al. introduced circulating miRNA-92a as a novel BC biomarker. The authors proposed that reduced levels of miRNA-92a and amplified levels of miRNA-21 in tissue and serum samples of BC were related to tumor size and a positive lymph node status compared to clinicopathologic data of the BC patients. In addition, miRNA-1, miRNA-92a, miRNA-133a, and miRNA-133b have been identified as the critical diagnostic biomarkers. These miRNAs were up-regulated and successfully validated in both serum and tumors of BC patients [41]. Chan et al. also reported six tumor-derived miRNAs as novel markers in BC patients, including upregulation of miRNA-720, miRNA-1274b, and miRNA1260 (as oncomirs) and down-regulation of miRNA-30c, miRNA-376c and miRNA-4324 (as tumor suppressors) in BC tumors [42]. Sochor et al. suggested that the surgery had tremendous impact on three sera oncomir levels (miR-155, miR-181b, and miR-24), whilst adjuvant therapy (chemo or radiotherapy after surgery) remarkably reduced all of the four oncomirs (miR-155, miR-181b, miR-24 & miR-19a) studied. The authors proposed that the oncomir levels are naturally abundant in the blood serum of early-stage breast cancer (EBC) patients as compared to healthy controls, so this feature can be used to identify EBC risk [43]. Erbes et al. assessed urinary BC-related miRNA expression levels, including miRNA-21, miRNA-34a, miRNA-125b, miRNA-155, miRNA-195, miRNA-200b, miRNA-200c, miRNA-375, miRNA-451 in 24 untreated, primary BC patients and 24 healthy controls. Using non-invasive method for BC detection, they indicated that four miRNAs (miR-21, miR-125b, miR-451 and miR-155) were significantly regulated in BC patients compared to healthy controls. In fact, the levels of miRNA-155 were higher, while the levels of miRNA-21, mi-RNA-125b and miRNA-451 were lower in BC patients [44]. Shimomura et al. comprehensively evaluated the expression profiles of serum miRNA in a training cohort (including non-cancer controls, other cancers and BC) and a test cohort (including non-cancer controls and BC) to determine novel miRNAs that can be used to diagnosis EBC and distinguish BC from other cancers, such as pancreatic, biliary tract, and benign prostate cancer. In their study, they suggest measurement of serum miRNA-1246, miRNA-1307-3p, miRNA-4634, miRNA-6861-5p and miRNA-6875-5p in combination could be useful [45]. Moreover, in a two-stage experimental design study by Tsai et al., the miRNA expression profiles in four groups with a differential age-onset, consisting of very young BC (n=15, ≤35 y), young BC (n=37, 36-40 y), premenopausal BC (n= 47, 41-50 y), postmenopausal BC (n=46, >50 y), was detected in Asian BC. The authors measured 85 in 145 BC samples. For all patients, two miRNAs (miRNA-335 and miRNA-145) were down-regulated and four miRNAs (miRNA-21, miRNA-200a, miRNA-200c, and miRNA-141) were up-regulated. Also, in very young BC, the expression of three miRNAs (miRNA-96, miRNA-182, and miRNA-183) and eight miRNAs (miRNA-10a, miRNA-10b, miRNA-125b, miRNA-127-3p, miRNA-130a, miRNA-143, miRNA-19, and miRNA-320) were down- and up-regulated, respectively. Furthermore, this study showed that miRNA-30b, miRNA-30d, and miRNA-149 were up-regulated and miRNA-487b, miRNA-548c-5p, and miRNA-181d were down-regulated in young BC. So far, only down-regulation of miRNA-206 has been detected in premenopausal BC, but no alteration has been observed in postmenopausal BC (Table 1) [46].
It seems that circulating miRNAs can be considered key biomarkers in the reduction of BC prevalence by enhancing the diagnosis, prognosis, prediction and the clinical management of patients and their relatives. Accumulative evidence in recent years has highlighted the significant role of circulating nucleic acids in the blood, especially miRNAs, as early detection markers [81,82], and even for monitoring patient responses to various therapies for several unrelated diseases [83-86]. Moreover, assessment of miRNAs expression levels in other body fluids such as urine, is another non-invasive method for the detection of BC that works with miRNA markers [84].
There is clear evidence of the prominent role of DNA methylation and miRNAs in cancer initiation, progression and even prevention [87]. It seems that in the epigenetic mechanisms, there is a bilateral relationship between DNA methylation pattern and miRNA expression profiles [88]. This mutual relationship can contribute to the diagnosis, prognosis and characterization of tumor tissue origination, metastatic capability and tumorigenesis potential in early stages of cancer [89]. In fact, by two major processes miRNAs can regulate DNA methylation in human cancers: 1) by modifying DNMTs and 2) by adjusting methylation of some crucial proteins, such as methyl CpG binding protein 2 (MeCP2) and methyl-CpG binding domain proteins 2 and 4 (MBD2 and MBD4) [87]. In BC, miRNA-221 directly targets DNMT3b [88], miRNA-143 negatively correlate with DNMT3a [90], and miRNA-29b object DNMT3a and 3b [91].
DNA methylation can also cause down-regulation of tumor-suppressor miRNAs by hyper-methylation and up-regulation onco-miRNAs by hypo-methylation [92]. The majority of the epigenetic studies have focused on quelling of tumor-suppressor miRNAs by hyper-methylation of CGI promoters in a variety of human cancers [93]. Saito et al. first indicated that miRNA-127 acts as a tumor suppressor and up-regulates via downregulation of its target; the proto-oncogene B-cell lymphoma 6 (BCL6). Moreover, aberrant methylation of the CGI in miRNA gene promoters in BC cells includes miRNA-125b (target gene: ETS1), miRNA-148a (target gene: TGIF2), miRNA-200 family (target gene: ZEB1 & ZEB2) and miRNA-335 (target gene: SOX4 & TNC). In addition, some miRNA genes are hyper-methylated in the CGI shore region, including miRNA-9, miRNA-149, miRNA-210, miRNA-212, miRNA-328, miRNA-503, miRNA-1224, miRNA-1227 and miRNA-1229 [87,94,95]. Figure 4 illustrates the pivotal role of aberrant DNA methylation and mutual association between miRNA and DNA methylation in human cancer.
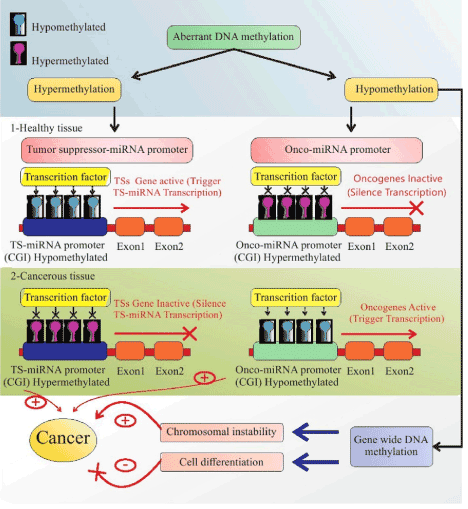
Figure 4. Amplification roles of miRNAs and DNA methylation on each other in healthy and cancerous cells. A pivotal role of aberrant DNA methylation in 1. Healthy tissue: Hypo-methylation of the CGI of the TS-miRNA promoter and hyper-methylation of the CGI of the onco-miRNA promoter causes activation of TSGs and inactivation of oncogenes, respectively. DNA methylation is adjusted by miRNA via a DNMT complex or major proteins that engaged in methylation. 2. In cancerous tissues: The promoters of TSGs are down-regulated via their hyper-methylation and lead to proliferation of cells and tumorigenesis. Oncogenes are up-regulated by hypo-methylation, then exert their tumorigenesis roles. Genome-wide (GW) DNA methylation followed by GW hypo-methylation leads to chromosomal instability and cell differentiation, resulting in tumorigenesis and its inhibition, respectively. TSGs: Tumor Suppressor Genes; GW: Genome Wide
Essential and non-essential nutrients in BC
The role of some lifestyle factors, including BMI (overweight and obesity), habits, physical activity, smoking, and breastfeeding on BC diagnosis and prognosis have been assessed by various studies and the relationship between dietary patterns and nutrients with BC is inconsistent [96,97]. However, it has been suggested that one-third of all cancers in the USA are preventable by a change of diet and certain nutrients [98], and there are some studies showing that BC initiation and progression is at least in part, influenced by the diet. Essential nutrients, such as Folate, vitamin B12, vitamin D and non-essential nutrients like resveratrol, curcumin, tea phenols, genistein, sulforaphane, methionine, choline, and betaine can affect DNMT, DNA methylation, miRNAs oncogenic and tumor-suppressor miRNAs [99-101], highlighting a possible role of nutrigenomics in the epigenetic mechanisms of BC. The evidence shows that a healthy food plan specified by a high intake of vegetables, fruits, whole grains and legumes like beans, peas, lentils, and poultry, as well as low-fat and high-fiber dietary products, may impact on BC progression [101,102], whilst an unhealthy diet including a high intake of red or processed meat, high fat (saturated and trans fatty acid) dairy products and low intake of omega-3 fatty acids, refined grains and sugars, low intake of natural antioxidants, and fiber, can trigger the inflammation process, possibly enhancing the risk of BC [96,102]. Using nutritional genomics, we can elucidate if and how these different nutrients modify protein and gene expression leading to altered cellular function. Nutrition components can either act as carcinogenesis risk factors or show anti-cancer functions in cells and participate in initiation, progression and metastasis, or prevention and treatment of cancer cells [103]. The number of studies, which intend to clarify the effects of nutrients on the regulation of epigenetic mechanisms involved in BC prevention, is increasing and a few of them have been recapped in Table 3.
Table 3. Summary of nutrigenomics and BC studies
References |
Nutrient components |
Major finding |
Roystone et al. (2018) [104] |
5.0 µM SFN, 1.0 µM WA or both. |
- In MCF-7 cells: SFN or WA reduced transition into the G2/M phase, whereas the combination of them arrested cells at the G1 phase.
- In MDA-MB-231 cells: SFN+WA showed a decline in transition into S phase, and SFN consider to be more effective than WA.
- CCND1 and CDK4 (promote transition from G1 to S phase) expression reduced with the SFN and WA alone and also pRB (G1 checkpoint) decreased, while the combination of SFN+WN was not most effective in both cell lines BC cells.
- Up-regulation of E2F CDK4 (promote transition from G1 to S phase) in both cell lines was most effective by SFN+WA.
- WA increased HAT activity in both cell lines but it was significant in MDA-MB-231 cells, in comparison with SFA+WA and SFA.
- SFA+WA significantly increased global methylation in both MCF-7 and MDA-MB-231 cells.
- P21expression increased in MDA-MB-231 cells, and P-53 reduced by SFN+WA. Whereas, it was observed mild go up in P21 and P53 in MCF-7 cells by SFN or WA and both of them.
- SFN or WA alone, or in combination with WA mainly augmented the surfaces of H3K4Me3 at the p21 promoter, showing that these nutrient compounds positively modulate the epigenome at the promoter region. |
Zhu et al. (2012) [105] |
- supplementing with capsule twice daily for 12 weeks containing 1 of 3 preparations:
1-placebo, 2-P. cuspidatum with 5 mg trans-resveratrol, 3-P. cuspidatum with 50 mg of trans resveratrol |
- RASSF-1α (BC related gene) methylation declined with increasing levels of serum trans-resveratrol. |
Qin et al. (2014) [106] |
- Rats were randomly divided in 4 groups:
1- Control group,
2- Low dose resveratrol (lo; 5 mg/kg/day),
3- High dose resveratrol (hi; 25 mg/kg/day),
4- Resveratrol in a Harlan Teklad prepared diet, or 0.1 mg/kg/wk 5-aza-2 deoxycytidine by intraperitoneal injection, |
- Hi resveratrol and Aza provoked a differential impact on DNMT3b expression in tumor in comparison with normal tissue. DNMT3b reduced in tumor but up-surged in normal tissue.
- Lo but not hi resveratrol augmented miR10a and miR10b expression in tumor tissue.
- Hi dose augmented miRNA-21, -129, -204, and -489 and reduced the same miRs in normal tissue.
- It was observed contradict association between DNMT3b and miRNA-129, -204, and -489 in normal and/or tumor tissue. |
D. Adams et al. (2018) [107] |
- |
- Four miRNAs, including miRNA-22-3p, miRNA-122-5p, miRNA-126-3p, miRNA-150-5p have positive correlation with BMI, whilst miRNA-191-5p, miRNA-17-5p, miRNA-103a-3p, miRNA-93-5p have negative association with BMI in HOPE trial.
- Expression of miRNA-106b-5p, let-7b-5p, and miRNA-92a-3p decreased, whereas miRNA-27a-3p, miRNA-191-5p, miRNA-24-3p increased at the end of the six-month intervention by arm in the LEAN trial. |
Pietruszewska et al. (2015) (108) |
- MCF-7 and MDA-MB-23 cells cultured for 96 h with SFN at different concentrations. |
- In MCF-7 cells at 10 and 22 μM concentrations of SFN: PTEN and RARbeta2 promoter methylation decreased by 25 and 12 %, respectively, whilst PTEN mRNA upregulated by 32% in 10 μM and 82% in 22 μM and RARbeta2 expression increased 55–60%.
- In MDA-MB-231 cells at 10 and 46 μM concentrations of SFN: decreased in PTEN promoter methylation by 22 and 31%, and PTEN mRNA expression increased by 72 and 101%, respectively.
- In MCF-7 cells, p21 expression increased by 2.5- and 1.8-fold, and p53 mRNA levels augmented by 66 and 38% at 10 and 22 μM SFN concentrations, respectively.
- In MDA-MB-231 cells which express mutant p53, SFN at 10 and 46 μM concentrations enhanced p21 expression by 3-fold and 55%, respectively.
- Although regulators of DNMT1, p21 and p53 over-expressed, there was no effect on DNMT1 suppression. |
Peng et al. (2017) [109] |
20 mg/ kg/day of 3,6-DHF |
- 3,6-DHF could reverse the global down-regulation of miR-34a in BC by regulating the miR-34a promoter methylation.
- 3,6-DHF up-lifted TET1 level in MDA-MB-231 cells, and had no influence on TET2 and TET3.
- 3,6-DHF prohibited DNMT1 activity, modified the disequilibrium of DNA methylation and demethylation status. |
Kumar et al. (2017) [110] |
- MCF-7 cells treated with curcumin at 0, 1, 3, 5, 10, 20, 30, 50, and 100 µM for different time periods, including 24, 48, and 72h. |
- Treated MCF-7 cells with 10 µM of curcumin for 72h reversed the GSTP1 promoter hyper-methylation and re-expressed GSTP1 protein. |
Pietruszewska et al. (2013) [111] |
- ER-positive MCF-7 and invasive ER-negative MDAMB-231 cells, treated with 1 mg/l in control and 4 mg/l and 8 mg/l in experiment groups. |
- In high dose administration of folic acid, down-regulation of tumor suppressor genes, including PTEN, APC and RARbeta2 and increasing DNA methylation in their promoters occurred in MCF-7 cells.
- Induced DNMT1 expression in MCF-7 cells in highest dose. |
SFN: Sulforaphane; WA: Withaferin A; CCND1: Cell cyclin D1; CDK4: Cyclin D kinase 4; pRB: phosphorylated retinoblastoma protein; HAT: Histone acetyltransferase activity; H3K4Me3: Trimethylated lysine 4 of histone; HOPE: Hormones and physical exercise; LEAN: Lifestyle, exercise, and nutrition; SFN: sulforaphane; 3,6-DHF: 3,6-dihydroxyflavone; TET: Ten-eleven translocation; GSTP1: Glutathione S-transferase pi 1.
Bioactive components and epigenetics in BC
Roystone et al. explored the effects of Withaferin A (WA, found in the Indian winter cherry) and sulforaphane (SFN, found in cruciferous vegetables) acting as a DNMT inhibitor and histone deacetylase (HDAC) inhibitor, respectively. WA and SFN play a role in the regulation of cell cycle, and epigenetic-modifying enzymes in MCF-7 and MDA-MB-231BC cell lines, highlighting the capability of WA and SFN in enhancement of apoptosis, cell death and reduction of malignant expression of certain genes in BC cells [104-112]. Administration of WA and SFN inhibited the cell cycle progression from S to G2 phase through down-regulation of cyclin D1 (CCND1), cyclin D kinase 4 (CDK4), phosphorylated retinoblastoma protein (pRB), and up-regulation of E2F mRNA. In addition, tumor suppressor p21 protein was increased in response to WA and SFN treatment, which may in part relate to the downregulation of pRB gene. In the same study, enhanced global methylation of MCF-7 and MDA-MB-231BC cell lines in BC cells was found [104]. Zhu et al. showed that trans-resveratrol can induce mammary promoter hyper-methylation in women at high risk for BC. The authors showed that two significant roles of trans-resveratrol include downregulation of DNMT [105] and cancer promoting prostaglandin (PG) E2 [113], as observed by in vitro investigations. It is also concluded that after increasing the Trans-isomer and the glucuronide metabolite predominate in the circulation throughout 12 week supplementation with trans-resveratrol, methylation of RASSF-1α which is known as a tumor suppressor of BC is decreased in addition to a reduction in PGE2 levels in breast cells [105]. Similarly, Qin et al. showed that resveratrol can reduce DNMT 1 and 3b expression in vitro and demethylate tumor suppressor RASSF-1α in individuals with a risk of BC development. In summary, they found that high doses of resveratrol (25 mg/kg/day) down-regulated DNMT 3b in tumor tissue compared to healthy tissues, while this effect was not observed for DNMT1. Additionally, tumor suppressive miRNAs: miRNA-21, miRNA-129, miRNA-204 and miRNA-489, were up-regulated more than two fold in tumor tissue and down-regulated two to 10 times in normal tissue, therefore negatively correlating with DNMT3b levels [106]. Pietruszewska et al. investigated the impact of sulforaphane (SFN) on the expression of DNA methylation-regulators in MCF-7 and MDA-MB-231 BC cells, including DNMT1, p53, and p21, as well as the methylation of PTEN and RARbeta2 tumor suppressor genes. They found that 22 µM or 10-46 µM of SFN best inhibited the growth of 50% of MCF-7 or MDA-MB-231 BC cells, and caused hypo-methylation of PTEN and RARbeta2 promoters, whilst their mRNA expression was up-regulated. Moreover, the expression of DNA methylation-regulators (p21 or p53) increased in both BC cell lines. Although the gene regulators of DNMT1, p21 and p53 were over-expressed in BC cell lines, no effect on DNMT1 suppression was observed [108]. However, in another study by this group observed an inverse relationship between p21 and DNMT1 expression because p21 competes with DNMT1 for the same binding site on a proliferating cell nuclear antigen (PCNA) during DNA replication [114]. Peng et al. suggest that 3,6- dihydroxyflavone (3,6-DHF) enhances miRNA-34a expression significantly in BC cells, and this component acts as a DNMT1 inhibitor in MDA-MB-231 cells [115]. More recently, 3,6-DHF has been found to up-regulate TSGs, such as miRNA-34a, through methylation of relevant promoters by suppressing DNMT1 during tumorigenesis. The level of ten-eleven translocation (TET) and 5-hydroxymethylcytosine (5hmc) are usually decreased in cancer cells, but administration of 3,6-DHF was found to increase the level of TET1 and 5hmc in MDA-MB-231 cells. Finally, because TET1 inhibition by siRNA was able to counteract the impact of 3,6-DHF on miRNA-34a promoter-demethylation and subsequent up-regulation, it has been proposed that an up-ward trend in TET1 expression through 3,6-DHF supplementation could be a nutritional measure to prevent BC [109]. A study by Kumar et al. investigated the effect of curcumin administration on the methylation status of glutathione S-transferase pi 1 (GSTP1) gene (a TSG) in MCF-7 BC cell line. While the ideal dose (IC50) dose of curcumin was 20µM, the non-toxic concentration of curcumin (10µM) reversed the hyper-methylation of the GSTP1 promoter and induce re-expression of GSTP1 protein [110]. Importantly, the authors noted that less than 3µM curcumin does not change the promoter methylation pattern of GSTP1 and greater than the IC50 and curcumin reinstates hyper-methylation of GSTP1 [110].
Essential components, BMI and epigenetics in BC
Adams et al. found a positive correlation between blood serum miRNA expression and the BMI of BC survivors, in the Hormones and Physical Exercise (HOPE) trial. In addition, the expression of 35 miRNAs were analyzed after a 6-month weight-loss trial (Lifestyle, Exercise, and Nutrition; LEAN). The HOPE trial reported that eight novel miRNAs, including miRNA-22-3p, miRNA-122-5p, miRNA-126-3p, miRNA-150-5p positively correlated with BMI, while miRNA-191-5p, miRNA-17-5p, miRNA-103a-3p, miRNA-93-5p negatively correlated. In the LEAN trial, miRNA-106b-5p, let-7b-5p, and miRNA-92a-3p were down-regulated and miRNA-27a-3p, miRNA-191-5p, miRNA-24-3p up-regulated after the 6 months of weight loss [107]. Pietruszewska et al. assessed the impact of 1, 4, and 8 mg/l folic acid on DNA methylation of PTEN, APC and RARbeta2 tumor suppressor genes in MCF-7 and MDA-MB-231 BC cells. They found that by increasing the concentration of folic acid, DNA methylation of these tumor suppressor gene promoters increased, transcriptional activities of these genes was impaired, and dose-dependent down-regulation of tumor suppressor genes occurred. These effects were more tangible in non-invasive MCF-7 cells as well as they indicated 30% up-regulation of DNMT1 expression at 8 mg/l dose [111].
The impact of nutrigenomics on cancer is still unclear and it needs further research to determine the exact mechanisms that food components use to influence cancer processes. Thus, investigating the processes utilized by dietary components in BC development is an important future direction for nutrigenomics. Evidence shows that the epigenetic impact of dietary compounds can intervene in various cell functions, such as proliferation and differentiation, cell cycle regulation, apoptosis, metastasis and angiogenesis by modifying miRNA expression and DNA methylation. Thus, it seems that nutrigenomics may provide new insight on treatment and prevention strategies to many diseases, including cancer.
Hundreds of abnormal gene methylation patterns, either hyper-methylation of TSGs or hypo-methylation of oncogenes, have been identified in BC. The involved genes can differ from one person to another according to many factors including age, ethnicity and family history. Epigenetic pathways, such as aberrant DNA methylation and improper expression of miRNAs, have particularly profound effects on initiation of BC and participate in BC metastasis, cell proliferation, division and angiogenesis. Nowadays, the most important question is whether it is possible to control and handle the epigenetic mechanisms by producing some nutraceutical or pharmaceutical substances. In other words, if the underlying cause of BC is dysregulation of tumor suppressor miRNAs or onco-miRNA secretion, is it possible to control BC by producing drugs based on miRNAs or natural products? Another avenue of research yet to be explored is the possible role of other small non-coding RNAs, including snRNA, snoRNA and siRNA in treatment and diagnosis of the onset of BC, especially in individuals with a high risk for BC development. This would lead on to testing the effects of food components in regulating the expression or function of these small non-coding RNAs. The most pressing area for nutrigenomic research in relation to BC, is in improving the diagnosis and treatment methodologies, especially in the early stage of the disease.
Assessment of circulating nucleic acids like miRNA, mRNA and CF-DNA in body fluids, such as saliva, bronchial aspirates, sera, plasma, urine, and even in stool could be a modern and noninvasive method for monitoring the occurrence or development of cancer, early detection and its recurrence. Hence, understanding the mechanisms of nutrigenomics in relation to epigenetics and disease has the potential to revolutionize BC diagnosis methods, prevention and treatment therapies. Particular focus on diagnosis and treatment at the onset of BC, and in patients most at risk, will be most effective in fighting the disease.
Authors are thankful to the Department of Pharmaceutical Sciences, Division Pharmacology, Faculty of Science Utrecht University and School of Nutrition and Food Science, Tabriz University of Medical Sciences.
RH designed the study, carried out the study and prepared the first draft of manuscript. MM and SV designed the study, conceived the study and edited the manuscript. GF, commented on study design and critically editing the manuscript. All authors read and approved the final manuscript.
All authors have no conflict of interest to report.
- Bray F, Ferlay J, Soerjomataram I, Siegel R, Torre L, et al. (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin.
- Flister M, Bergom C (2018) Genetic modifiers of the breast tumor microenvironment. Trends in Cancer 4: 429-444.
- Economopoulou P, Dimitriadis G, Psyrri A (2014) Beyond BRCA: New hereditary breast cancer susceptibility genes. Cancer Treatment Reviews.
- Greenman Ch, Stephens Ph, Smith R, Dalgliesh G, Hunter Ch, et al. (2007) Patterns of somatic mutation in human cancer genomes. Nature 446: 153-158.
- Tehranifar P, Wu HC, McDonald JA, Jasmine F, Santella RM GI, et al. (2018) Maternal cigarette smoking during pregnancy and offspring DNA methylation in midlife. Epigenetics 13: 129-134.
- Mahmood N, Rabbani SH (2017) DNA methylation and breast cancer: Mechanistic and therapeutic applications. Trends in Cancer Research: 12.
- Győrffy B, Hatzis Ch, Sanft T, Hofstatter E, Aktas B, et al. (2015) Multigene prognostic tests in breast cancer: past, present, future. Breast Cancer Research 17: 11.
- Wise IA, Charchar FJ (2016) Epigenetic modifications in essential hypertension. Int J Mol Sci 17: 451-465.
- Braicu C, Mehterov N, Vladimirov B, Sarafian V, Nabavi S, et al. (2017) Nutrigenomics in cancer: revisiting the effects of natural compounds. Seminars in Cancer Biology.
- Sharma P, Dwivedi Sh (2017) Nutrigenomics and nutrigenetics: New insight in disease prevention and cure. Ind J Clin Biochem 32: 371-373.
- Virmani A, Pinto L, Binienda Z (2013) Food, nutrigenomics, and neurodegeneration—Neuroprotection by What You Eat. Mol Neurobiol 48: 353-362.
- Kang J (2013) Nutrigenomics and cancer therapy. J Nutrigenet Nutrigenomics 6: 1-2.
- Corcuff JB, Merched A (2016) Nutrigenomics and nutrigenetics: The basis of molecular nutrition. Molecular Basis of Nutrition and Aging.
- Nepomuceno JC (2013) Nutrigenomics and Cancer prevention. Cancer Treatment 17: 391-3416.
- Piątek W, Milewska M, Krasowska U, Terlikowska KM (2018) Nutrigenomics: a new approach for cancer prevention and treatment. Progress in Health Sciences 8: 155-161.
- Dates C, Tollefsbol T (2018) Transforming Cancer epigenetics using nutritive approaches and non-coding RNAs. Curr Cancer Drug Target 18: 32-38.
- Anand P, Kunnumakkara AB, Sundaram C, Harikumar KB, Tharakan ST, et al. (2008) Cancer is a preventable disease that requires major lifestyle changes. . Pharm Res 25: 2097-2116.
- Mahmood N, Arakelian A, Muller W, Szyf M, Rabbani Sh (2018) Pharmacologic targeting of DNA methylation blocks breast cancer growth and metastasis. Cancer Research 78: 1388.
- Prinz-Langenohl R, Fohr I, Pietrzik K (2001) Beneficial role for folate in the prevention of colorectal and breast cancer. European Journal of Nutrition 40: 98-105.
- Lim DHK, Maher ER (2010) DNA methylation: a form of epigenetic control of gene expression. The Obstetrician & Gynaecologist 12: 37-42.
- Varela-Rey M, Woodhoo A, Martinez-Chantar ML, Mato J, Lu Sh (2013) Alcohol, DNA Methylation, and Cancer. Alcohol Research 35: 25-35.
- Rodríguez-Paredes M, Esteller M (2011) Cancer epigenetics reaches mainstream oncology. Nature Medicine 17: 330-339.
- Portela A, Esteller M (2010) Epigenetic modifcations and human disease. Nature Biotechnology 28: 1057-1068.
- Moore LD, Le T, Fan G (2014) DNA methylation and its basic function. Neuropsychopharmacology 38: 23-38.
- Veeck J, Esteller M (2010) Breast cancer epigenetics: From DNA methylation to microRNAs. J Mammary Gland Biol Neoplasia 15: 5-17.
- Messerschmidt DM, Knowles BB, Solter D (2014) DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes and Development 28: 812-828.
- Pietschmann A, Mehdipour P, Atri M, Hofmann W, Hosseini-Asl SS, et al. (2005) Mutation analysis of BRCA1 and BRCA2 genes in Iranian high risk breast cancer families. Journal of Cancer Research and Clinical Oncology 131: 552-558.
- Croes L, de Beeck KO, Pauwels P, Berghe WV, Peeters M et al. (2017) DFNA5 promoter methylation a marker for breast tumorigenesis. Oncotarget 8: 31948-31958.
- Kim MS, Lee J, Oh T, Moon Y, Chang E et al. (2012) Genome-wide identifcation of OTP gene as a novel methylation marker of breast cancer. Oncology Reports 27: 1681-1688.
- DeRoo L, Bolick S, Xu Z, Umbach D, Shore D, et al. (2014) Global DNA methylation and one-carbon metabolism gene polymorphisms and the risk of breast cancer in the sister Study. Carcinogenesis 35: 333-338.
- Chimonidou M, Tzitzira A, Strati A, Sotiropoulou G, Sfikas C, et al. (2013) CST6 promoter methylation in circulating cell-free DNA of breast cancer patients. Clinical Biochemistery 46: 235-240.
- Rauscher G, Kresovich J, Poulin M, Yan L, Macias V, et al. (2015) Exploring DNA methylation changes in promoter, intragenic, and intergenic regions as early and late events in breast cancer formation. BMC Cancer 15: 816.
- Stefanssona O, Morana S, Gomeza A, Sayolsa S, Arribas-Jorbaa C, et al. (2015) A DNA methylation-based definition of biologically distinct breast cancer subtypes. Molecular Oncology 9: 555-568.
- Tian HP, Lun ShM, Huang HJ, He R, Kong PZh, et al. (2015) DNA methylation affects the SP1-regulated transcription of FOXF2 in breast cancer cells. The Journal of Biological Chemistry 290: 19173-19183.
- Wu HCh, Southey M, Hibshoosh H, Santella R, Terry MB (2017) DNA methylation in breast tumor from high-risk women in the breast cancer family registry. Anticancer Research 37: 659-664.
- Dehbid M, Aleyasin SA, Vaziri H (2016) Evaluation of DNA Methylation of MAP9 gene in breast cancer as epigenetic biomarker. Molecular Biomarkers & Diagnosis 8: 1-6.
- Joo J, Dowty J, Milne R, Wong EM, Dugué PA, et al. (2018) Heritable DNA methylation marks associated with susceptibility to breast cancer. Nature Communications 9: 867.
- Fleischer Th, Tekpli X, Mathelier A, Wang Sh, Nebdal D, et al. (2017) DNA methylation at enhancers identifies distinct breast cancer lineages. Nature Communications 8: 1379.
- Ng E, Li R, Shin V, Jin HCh, Leung C, et al. (2013) Circulating microRNAs as specific biomarkers for breast cancer detection. PLoS One 8: e53141.
- Cuk K, Zucknick M, Heil J, Madhavan D, Schott S, et al. (2013) Circulating microRNAs in plasma as early detection markers for breast cancer. International Journal of Cancer 132: 1602-1612.
- Si H, Sun X, Chen Y, Cao Y, Chen Sh, et al. (2013) Circulating microRNA-92a and microRNA-21 as novel minimally invasive biomarkers for primary breast cancer. J Cancer Res Clin Oncol 139: 223-229.
- Chan M, Liaw ChS, Ji Sh M, Tan HH, Wong ChY, et al. (2013) Identification of circulating microRNA signatures for breast cancer detection. Clinical Cancer Research.
- Sochor M, Basova P, Pesta M, Dusilkova N, Bartos J, et al. (2014) Oncogenic MicroRNAs: miR-155, miR-19a, miR-181b, and miR-24 enable monitoring of early breast cancer in serum. BMC Cancer 14: 448.
- Erbes Th, Hirschfeld M, Rücker G, Jaeger M, Boas J, et al. (2015) Feasibility of urinary microRNA detection in breast cancer patients and its potential as an innovative non-invasive biomarker. BMC Cancer 15: 193.
- Shimomura A, Shiino Sh, Kawauchi J, Takizawa S, Sakamoto H, et al. (2016) Novel combination of serum microRNA for detecting breast cancer in the early stage. Cancer Science 107: 326-334.
- Tsai HP, Huang ShF, Li ChF, Chien HT, Chen ShCh (2018) Differential microRNA expression in breast cancer with different onset age. PLoS One 13: 1-19.
- Hayes J, Peruzzi PP, Lawler S (2014) MicroRNAs in cancer: biomarkers, functions and therapy. Cell press 20: 460-469.
- Luceri C, Bigagli E, Pitozzi V, Giovannelli L (2015) A nutrigenomics approach for the study of anti‑aging interventions: olive oil phenols and the modulation of gene and microRNA expression profles in mouse brain. European Journal of Nutrition: 1-13.
- Denli AM, Tops BB, Plasterk RH, Ketting R, Hannon G (2004) Processing of primary microRNAs by the Microprocessor complex. Nature 432: 231-235.
- Lee Y, Ahn C, Han J, Choi H, Kim J, et al. (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415-419.
- Takahashi RU, Miyazaki H, Ochiya T (2015) The roles of microRNAs in breast cancer. Cancers 7: 598-616.
- Bertoli G, Cava C, Castiglioni I (2015) MicroRNAs: New biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics 5: 1122-1143.
- Goh J, Loo S, Datta A, Siveen K, Yap W, et al. (2016) microRNAs in breast cancer: regulatory roles governing the hallmarks of cancer. Biol Rev 91: 409-428.
- Arefhosseini SR, Ebrahimi-Mamaeghani M, Mohammadi S (2014) MicroRNAs regulation by nutrients, the new ray of hope in obesity related glucose and lipid metabolic disorders. Metabolic Syndrome 3: 1-12.
- Ruan K, Fang X, Ouyang G (2009) MicroRNAs: Novel regulators in the hallmarks of human cancer. Cancer Letter 285: 116-126.
- Jit SK (2012) Circulating microRNAs as biomarkers, therapeutic targets, and signaling molecules. Sensors (Basel) 12: 3359-3369.
- lcochova H, Hezova R, Stanik M, Sloby O (2014) Urine microRNAs as potential noninvasive biomarkers in urologic cancers. Urol Onco 32: 41-49.
- Shen J, Stass SA, Jiang F (2013) MicroRNAs as potential biomarkers in human solid tumors. Cancer Lett 329: 125-136.
- Trang Ph, Weidhaas J, Slack FJ (2017) MicroRNAs and cancer. The Molecular Basis of Human Cancer.
- Oliveto S, Mancino M, Manfrini N, Biffo S (2017) Role of microRNAs in translation regulation and cancer. World J Biol Chem 8: 45-56.
- Schooneveld EV, Wildiers H, Vergote I, Vermeulen PB, Dirix LY, et al. (2015) Dysregulation of microRNAs in breast cancer and their potential role as prognostic and predictive biomarkers in patient management. Breast Cancer Research 17: 21.
- Ahmad J, Hasnain S, Siddiqui M, Ahamed M, Musarrat J, et al. (2013) MicroRNA in carcinogenesis & cancer diagnostics: A new paradigm. Indian J Med Res 137: 680-694.
- Sandhu SK, Fassan M, Volinia S, Lovat F, Balatti V, et al. (2013) B-cell malignancies in microRNA Eμ-miR-17~92 transgenic mice. Proc Natl Acad Sci USA 110: 18208-1813.
- Wu H, Wang J, Ma H, Xiao Z, Dong X (2017) MicroRNA-21 inhibits mitochondria-mediated apoptosis in keloid. Oncotarget.
- Hou X, Zhang M, Qiao H (2015) Diagnostic significance of miR-106a in gastric cancer. Int J Clin Exp Pathol 8: 13096-13101.
- Ma L, Teruya-Feldstein J, Weinberg R (2007) Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449: 682-688.
- Nagpal N, Ahmad HM, Chameettachal S, Sundar D, Ghosh S, et al. (2015) HIF-inducible miR-191 promotes migration in breast cancer through complex regulation of TGFβ-signaling in hypoxic microenvironment. Sci Rep 5: 9650.
- Wang X, Cao L, Wang Y, Wang X, Liu N, You Y (2012) Regulation of let-7 and its target oncogenes. Oncol Lett 3: 955-960.
- Reid G (2015) MicroRNAs in mesothelioma: from tumour suppressors and biomarkers to therapeutic targets. J Thorac Dis 7: 1031-1040.
- Mizuno K, Seki N, Mataki H, Matsushita R, Kamikawaji K, et al. (2016) Tumor-suppressive microRNA-29 family inhibits cancer cell migration and invasion directly targeting LOXL2 in lung squamous cell carcinoma. International journal of Oncology 48: 450-460.
- Ito Y, Inoue A, Seers T, Hato Y, Igarashi A, Toyama T (2017) Identification of targets of tumor suppressor microRNA-34a using a reporter library system. PNAS 114: 3927-3932.
- Tomasetti M, Monaco F, Manzella N, Rohlena J, Rohlenova K, et al. (2016) MicroRNA-126 induces autophagy by altering cell metabolism in malignant mesothelioma. Oncotarget 7: 36338-36352.
- Guo Y, W F, Chen H, Shang C, Zhong M (2012) miR-24 functions as a tumor suppressor in Hep2 laryngeal carcinoma cells partly through down-regulation of the S100A8 protein. Oncol Rep 27: 1097-1103.
- Zhao G, Liu L, Zhao T, Jin S, Jiang S, Cao S (2015) Upregulation of miR-24 promotes cell proliferation by targeting NAIF1 in non-small cell lung cancer. Tumour Biol 36: 3692-3701.
- Sun YM, Lin KY, Chen YQ (2013) Diverse functions of miR-125 family in different cell contexts. J Hematol Oncol 6: 6.
- Li CL, Nie H, Wang M, Su LP, Li JF, et al. (2012) microRNA-155 is downregulated in gastric cancer cells and involved in cell metastasis. Oncol Rep 27: 1960-1966.
- Garofalo M, Quintavalle C, Romano G, Croce M, Condorelli G (2012) miR221/222 in cancer: their role in tumor progression and response to therapy. Curr Mol Med 12: 27-33.
- Jafari N, Peeri Dogaheh H, Bohlooli Sh, G Oyong G, Shirzad Z, et al. (2013) Expression levels of microRNA machinery components Drosha, Dicer and DGCR8 in human (AGS, HepG2, and KEYSE-30) cancer cell lines. International Journal of Clinical and Experimental Medicine 6: 269-274.
- Xin H, Li X, Yang B, Zhang L, Han Z, et al. (2014) Blood-based multiple-microRNA assay displays a better diagnostic performance than single-microRNA assay in the diagnosis of breast tumor. Tumour Biol 35: 12635-12645.
- Hemmatzadeh M, Mohammadi H, Jadidi-Niaragh F, Asghari F, Yousefi M (2016) The role of oncomirs in the pathogenesis and treatment of breast cancer. Biomedicine & Pharmacotherapy 78: 129-139.
- Chen X, Ba Y, Ma L, Cai X, Yin Y, et al. (2008) Characterization of microRNAs in serum: anovel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 18: 997-1006.
- Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, et al. (2008) Serum microRNAsare promising novel biomarkers. PLoS ONE 3: e3148.
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, et al. (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl AcadSci USA 105: 10513-10518.
- Hamam R, Hamam D, Alsaleh KhA, Kassem M, Zaher W, et al. (2017) Circulating microRNAs in breast cancer: novel diagnostic and prognostic biomarkers. Cell Death and Disease 8: e3045.
- Guay C, Regazzi R (2013) Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat Rev Endocrinol 9: 513-521.
- Gandhi R, Healy B, Gholipour T, Egorova S, Musallam A, et al. (2013) CirculatingmicroRNAs as biomarkers for disease staging in multiple sclerosis. Neurol 73: 729-740.
- Wang S, Wu W, Claret F (2017) Mutual regulation of microRNAs and DNA methylation in human cancers. Epigenetics 12: 187-197.
- Roscigno G, Quintavalle C, Donnarumma E, Puoti I, Diaz-Lagares A, et al. (2016) MiR-221 promotes stemness of breast cancer cells by targeting DNMT3b. Oncotarget 7: 580-592.
- Holubekova V, Mendelova A, Jasek K, Mersakova S, Zubor P, et al. (2017) Epigenetic regulation by DNA methylation and miRNA molecules in cancer. Future Oncol 13: 2217-2222.
- Ng E, Li R, Shin V, Siu J, Ma E, et al. (2014) MicroRNA-143 is downregulated in breast cancer and regulates DNA methyltransferases 3A in breast cancer cells. Tumor biology 35: 2591-2598.
- Starlard-Davenport A, Kutanzi K, Tryndyak V, Word B, Lyn-Cook B (2013) Restoration of the methylation status of hypermethylated gene promoters by microRNA‑29b in human breast cancer: A novel epigenetic therapeutic approach. Carcinogenesis 12: 15.
- Pronina I, Loginova V, Burdennyy A, Fridman M, Senchenko V, et al. (2017) DNA methylation contributes to deregulation of 12 cancer-associated microRNAs and breast cancer progression. Gene 604: 1-8.
- Lujambio A, Calin G, Villanueva A, Ropero S, Sa ́nchez-Ce ́spedes M, et al. (2008) A microRNA DNA methylation signature for humancancer metastasis. Proc Natl Acad Sci USA 105: 13556-13561.
- Saito y, Liang G, Egger G, Friedman J, Chuang J, et al. (2006) Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 9(6).
- Suzuki H, Maruyama R, Yamamoto E, Kai M (2012) DNA methylation and microRNA dysregulation in cancer. Molecular Oncology 6: 567-578.
- Weigl J, Hauner H, Hauner D (2018) Can nutrition lower the risk of recurrence in breast cancer? Breast care 13: 86-91.
- Patterson RE, Cadmus LA, Emond JA, Pierce JP (2010) Physical activity, diet, adiposity and female breast cancer prognosis: a review of the epidemiologic literature. Maturitas 66: 5-15.
- Khan Sh, Aumsuwan P, Khan I, Walker L, Dasmahapatra A (2012) Epigenetic Events associated with breast cancer and their prevention by dietary components targeting the epigenome. Chem Res Toxicol 25: 61-73.
- Li E, Zhang Y (2014) DNA methylation in mammals. Cold Spring Harb Perspect Biol 6: a019133.
- Liu J, Ward RL (2010) Folate and one-carbon metabolism and its impact on aberrant DNA methylation in cancer. Adv Genet 71: 80-121.
- McKay J, Mathers J (2011) Diet induced epigenetic changes and their implications for health. Acta Physiol 202: 103-118.
- Seiler A, Chen M, Brown R, Fagundes Ch (2018) Obesity, dietary factors, nutrition, and breast cancer risk. Current Breast Cancer Reports: 1-16.
- Theodoratou E, Timofeeva M, Li X, Meng X, Ioannidis JPA (2017) Nature, nurture, and cancer risks: genetic and nutritional contribu-tions to cancer. Annu Rev Nutr 37: 293-320.
- Royston K, Paul B, Nozell S, Rajbhandari R, Tollefsbol T (2018) Withaferin A and sulforaphane regulate breast cancer cell cycle progression through epigenetic mechanisms. Experimental Cell Research: 1-8.
- Zhu W, Qin W, Zhang K, Rottinghaus G, Chen YCh, et al. (2012) Trans-resveratrol alters mammary promoter hypermethylation in women at increased risk for breast cancer. Nutrition and Cancer 64: 393-400.
- Qin w, Zhang K, Clarke K, Weiland T, Sauter E (2014) Methylation and miRNA effects of resveratrol on mammary tumors vs. normal tissue. Nutrition and Cancer 66: 270-277.
- Adams B, Arem H, Hubal M, Cartmel B, Li F, et al. (2018) Exercise and weight loss interventions and miRNA expression in women with breast cancer. Breast Cancer Research and Treatment: 1-13.
- Lubecka-Pietruszewska K, Kaufman-Szymczyk A, Stefanska B, Cebula-Obrzutb B, Smolewski P, et al. (2015) Sulforaphane Alone and in combination with clofarabine epigenetically regulates the expression of DNA methylation-silenced tumour suppressor genes in human breast cancer cells. J Nutrigenet Nutrigenomics 8: 91-101.
- Peng X, Chang H, Chen J, Zhang Q, et al. (2017) 3,6-Dihydroxyflavone regulates microRNA- 34a through DNA methylation. BMC Cancer 17: 619.
- Kumar U, Sharma U, Rathi G (2017) Reversal of hypermethylation and reactivation of glutathione S-transferase pi 1 gene by curcumin in breast cancer cell line. Tumor Biology: 1-8.
- Lubecka-Pietruszewska K, Kaufman-Szymczyk A, Stefanska B, Fabianowska-Majewsk K (2013) Folic acid enforces DNA methylation-mediated transcriptional silencing of PTEN, APC and RARbeta2 tumour suppressor genes in breast cancer. Biochemical and Biophysical Research Communications 430: 623-628.
- Royston K, Udayakumar N, Lewis K, Tollefsbol T (2017) A novel combination of withaferin A and sulforaphane inhibits epigenetic machinery, cellular viability and induces apoptosis of breast cancer cells. Int J Mol Sci 18: 1092.
- Masferrer JL, Leahy KM, Koki AT, Zweifel BS, et al. (2000) Antiangiogenic and antitumor activities of cyclooxygenase-2 inhibitors. Cancer Res 60: 1306-1311.
- Christman JK (2002) 5-Azacytidine and 5-aza-2’-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 21: 5483-5495.
- Peng X, Chang H, Gu Y, Chen J, Yi L, et al. (2015) 3,6-Dihydroxyflavone suppresses breast carcinogenesis by epigenetically regulating miR-34a and miR-21. Cancer Prev Res (Phila) 8: 509-517.

Figure 1. DNA methylation. DNA methylation is a post replicative process that results from transfer of a methyl (CH3) group to the 5-carbon of the cytosine (C5) ring by DNMTs, and is characterized as 5 methylcytosine (5-MC). Hyper-methylation of CGIs in gene promoters leads to alterations in chromatin structure and restricts access of TFs to the gene promoter, quelling the gene transcriptions and silencing of the gene. DNMT: DNA methyl transferase; TF: Transcription factor; CpG Islands

Figure 2. DNA methylation templates in normal and cancerous cells. (A) Repetitive Sequence: Hyper-methylation in repetitive sequences prohibit gene transcription in healthy cells, while reduction of 20 to 60 percent in the 5-methylcytosine in cancerous cells leads to deformation of chromatins and triggers gene transcription. (B) CpG Island Promoters: CpG island promoters are generally hypo-methylated and activate transcription in healthy cells. Conversely, CpG island promoters are hyper-methylated in cancerous cells, resulting in transcriptional inactivation. (C) CpG Island Shore: The mechanism of DNA methylation templates is the same as in B, except that the CpG island shore is located up-stream. (D) Gene Bodies: CpG methylation in gene bodies prohibit gene expression in healthy cells, but is reversed in cancer cells, resulting in initiation of transcription at several incorrect loci

Figure 3. The biogenesis of miRNA. Initially, primary transcription of miRNAs (pri-miRNAs), 1-3 Kb long, are produced by RNA polymerase II. Next, the pri-miRNAs are cleaved into stem-loop pre-miRNA (70-100 nucleotides long) by Drosha (RNA polymerase III) and double-stranded RNA binding protein DGCR8. Pre-miRNAs are transported to the cytoplasm by binding to Exportin-5 receptors (exporting-5-RanGTP) and then processed into a mature miRNA duplex by Dicer (RNase III enzyme), Trans Activating Response RNA-Binding Protein 2 (TARBP2) and AGO2 (DICER complex). The duplex strands are then separated: the complementary strand can be loaded in the RISC or is degraded, and the mature miRNA (the guide strand) is incorporated into the RNA-induced silencing complex (RISC) which includes the GW182 and Argonaut (AGO) proteins. The degree of homology of the miRNA “seed” to the 3′ UTR target sequence of the mRNA determines the mRNA degradation, translational inhibition, or transcriptional activation. Nonetheless, dysregulation of miRNAs expression is able to have a major role in the development of tumor progression, including tumor growth, invasion, proliferation, cell death and angiogenesis. An oncogenic miRNA is intended to repress the translation of a tumor suppressor gene, provoking tumorigenesis, angiogenesis, invasion and leading to tumor formation. Conversely, a tumor suppressor miRNA can prohibit the expression of oncogene. Pri-miRNA: primary miRNA; pre-miRNA: precursor miRNA

Figure 4. Amplification roles of miRNAs and DNA methylation on each other in healthy and cancerous cells. A pivotal role of aberrant DNA methylation in 1. Healthy tissue: Hypo-methylation of the CGI of the TS-miRNA promoter and hyper-methylation of the CGI of the onco-miRNA promoter causes activation of TSGs and inactivation of oncogenes, respectively. DNA methylation is adjusted by miRNA via a DNMT complex or major proteins that engaged in methylation. 2. In cancerous tissues: The promoters of TSGs are down-regulated via their hyper-methylation and lead to proliferation of cells and tumorigenesis. Oncogenes are up-regulated by hypo-methylation, then exert their tumorigenesis roles. Genome-wide (GW) DNA methylation followed by GW hypo-methylation leads to chromosomal instability and cell differentiation, resulting in tumorigenesis and its inhibition, respectively. TSGs: Tumor Suppressor Genes; GW: Genome Wide
Table 1. Summary of miRNAs, DNA methylation and BC studies
References |
Samples |
Methods |
Major finding |
Summary of DNA methylation studies |
Kim et al., 2012 [29] |
- BC cell lines: MCF-7 (HTB-22) and MDA-MB-231 (HTB-26).
-Fresh-frozen tissue specimens.
- Three candidate genes for methylation status in primary tumor, normal tissues, and paired adjacent normal appearing tissues (n=39):
LHX2, WT1 and OTP. |
- CpG microarray analysis: MeDIA + MBD2bt
- The hybridized images: analyzed by Agilent DNA microarray Scanner
- Bisulfite PCR and pyrosequencing primers: augment 3 to 5 CpG dinucleotide sites in the target areas of genes. |
- Aberrant hyper-methylation of genes in primary tumors (43.6% for LHX2, 89.7% for WT1 and 100% for OTP), whereas frequencies were invisible in normal tissues and intermediate in paired adjacent normal tissues.
- OTP gene most probably have a predominately role in breast carcinogenesis. |
DeRoo et al., 2014 [30] |
- A prospective cohort study among 50 884 (294 cases and 646 non-cases; ages: 35-75 years).
- LINE-1 methylation
was assessed in DNA from peripheral blood samples and blood clots. |
- Global methylation was gauged by pyrosequencing of the LINE-1 repetitive element. |
- Three SNP in genes are related to LINE-1 methylation: MTRR (rs10380), SLC19A1 (rs1051266), and MTHFR (rs1537514).
- MTHFR (rs1537514) was connected with BC risk.
- PON1 (rs757158) was associated with BC but not methylation. |
Chimonidou et al., 2013 [31] |
- 233 clinical samples:
a) One pilot testing group: 27 patients, 46 patients for verified metastasis & 37 healthy female (blood donors).
b) Independent cohort: 123 patients.
- The BC cell line: MCF 7 (positive control in MSP). |
-The methylation status of CST6 in circulating cfDNA was assessed by nested MSP. |
- CST6 promoter is crucially methylated in cfDNA of BC participants compared to healthy individuals.
- CST6 promoter methylation in cfDNA: recognized as a newfound plasma tumor biomarker for BC. |
Rauscher et al., 2015 [32] |
- BC patients: n = 129 (ages: 30-79).
- Control: 18 unaffected, cancer-free patients.
- The DNA regions studied were promoters, far-upstream regions, introns, and the LINE-1 and satellite 2 DNA repeats. |
- Pyrosequencing was conducted on bisulfite-treated DNA from FFPE |
- The majority of the assessed gene regions: hyper-methylated in cancer vs. adjacent tissue, except for TFF1 and MAGEA1 regions were highly hypo-methylated.
- Confirming frequent DNA methylation changes in invasive BC at a various genome location. |
Stefansson et al., 2015 [33] |
- Samples: 25 normal breast tissue (adjacent to tumor lesions of patients) and 350 primary invasive breast tumors. |
- DNA isolation: phenolchloroform/Proteinase-K method.
- DNA methylation analysis:Infinium HumanMethylation450 BeadChips.
- The PyroMark Q96 system for pyrosequencing was applied to
evaluate. |
-Discovered signatures related to two biologically distinct BC subtypes: Basal-like and Luminal-B.
-DNA methylation patterns linked to the Luminal-B subtype: CpG island promoter methylation.
-Basal-like tumors: hypo-methylation of gene body. |
Tian et al., 2015 [34] |
-BC cell lines: MCF-7, MDAMB-453, and MDA-MB-231 and the human mammary epithelial cell line: MCF-10A.
- 20 primary BC tissue specimens (invasive ductal carcinoma).
- DNA achieved from cultured cells or frozen tissue. |
-Using SssI, HhaI, and HpaII methylases or no enzyme (mock): Influence of CpG Island Methylation Status on FOXF2.
- Cell proliferation: 3-(4,5-dimethyl- thiazol-2-yl)- 2,5-diphenyl- tetrazolium bromide (MTT) assay
- Invasion and migration of breast cells: Matrigel-coated and uncoated Transwell, respectively. |
- CpG island methylation of the FOXF2 proximal promoter: engaged in the regulatory process of the subtype-specific expression of FOXF2 in BC cells.
-SP1: adjusted the transcriptional activity of FOXF2 by straight binding to the
proximal promoter region.
- Function and expression of FOXF2 in BC cells modulated through the composed impacts of DNA methylation and SP1 transcriptional regulation. |
Wu et al., 2017 [35] |
- 40 FFPE breast tumors and adjacent non-tumor tissues from BC cases (have family history of BC).
- The genes had selected from analysis of TCGA project. |
- DNA methylation: Illumina MiSEQ instrument (measuring DNA methylation in candidate genes.) |
-13 out of 40 genes were highly methylated.
- In regard to TCGA, methylation levels in SEPW1, PCDHGA4, CCDC36, C1orf14, RPTOR, C1orf114, ZNF454, USP44, CSMD3, PRKAR1B, SLC7A14, SOX2OT and RYR2 were remarkably observed in the breast tumor.
- 32% of tested genes: hyper-methylated in BCFR (36.1%: SEPW1, 10%: RYR2). |
Dehbid et al., 2016 [36] |
Samples: 30 healthy women and 30 BC women. |
- DNA isolated from 200 µl whole blood by MBST DNA extraction Kit.
- Detecting DNA with CPG island methylation by restriction endonuclease quantitative PCR method. |
- Hyper-methylation of MAP9 gene in BC can be used as molecular biomarker for BC diagnosis. |
Summary of miRNAs studies |
Ng et al., 2013 [39] |
- BC: n= 260 (clinically diagnosed),
- Normal subjects: n=170 (pathologically verified not to have BC and no history of other cancers).
- MiRNA profiling was proceeded in four groups: 1) BC plasma, 2) normal plasma, 3) tumor, and 4) adjacent non-tumor tissue. |
- Extraction total RNA from tissues: TRIzol reagent
- Extraction total RNA containing small RNA from 500 ml of plasma: Trizol LS reagent and miRNeasy Mini Kit.
-Plasma miRNA Profiling: using TaqMan Array Human miRNA.
- Plasma miRNA Quantitative PCR: SYBR green qRT-PCR assay |
- Crucial promotion of the miRNA- 16, miRNA-21, and miRNA-451 and remarkable decline of miRNA-145 in the plasma of BC patients.
- The composition of plasma miRNA-145 and miRNA-451 surfaces indicated the best markers for BC prediction. |
Cuk et al.,2013 [40] |
- Blood samples were prepared from 80 healthy and 127 female sporadic BC.
- Malignant tissue samples were provided from non-neoadjuvant BC patients during surgery. |
- Extraction total RNA from 400 ml of plasma: Trizol LS reagent and miRNeasy Mini Kit.
-Plasma miRNA Profiling: TLDA.
-Validation of chosen marker candidates: RT reactions were done by using TaqMan MicroRNA RT Kit and miRNA specific RT primers. |
- Four miRNAs (miRNA-148b, miRNA-376c, miRNA-409-3p and miRNA-801) significantly up-regulated in the plasma of BC patients.
- Based on ROC curve, only three miRNAs (miRNA-148b, miRNA-409-3p and miRNA-801) had an equal discriminatory power among BC samples and healthy controls. |
Si et al., 2013 [41] |
- Tumor tissues, paired NATs, and matching serum samples were collected from 48 newly diagnosed breast carcinomas.
- Other serum samples: 52 cases of BC and 20 of HC. |
- Total RNA isolation: The miRNeasy Mini Kit and mirVana PARIS Kit.
- MiRNA quantification: SYBR green qRT-PCR assay. |
- MiRNA-92a and miRNA-21 expression may be used to discriminate the BC patients from the HC.
- Circulating miRNA-92a has potential use as novel BC biomarker.
- Decreased levels of miRNA-92a and increased levels of miRNA-21 were connected with tumor size and a positive lymph node status in compared to clinicopathologic data of the BC patients. |
Chan et al., 2013 [42] |
- Extracted miRNA from paired fresh frozen BC tumors, normal tissue, and serum samples of patients (n=32).
- Serum samples from normal controls (n = 22).
- Additional serum samples from patients with BC (n = 132) and control serum samples (n = 101). |
- MiRNA extraction: From tissue or serum samples by miRVana or miRNeasy kits, respectively.
- Quality control of RNA from tissue samples: Agilent Bioanalyzer.
-Quality control of serum samples: singleplex LNA RT-PCR. |
- Only 7 miRNAs were upregulated in both serum and tumors.
- MiRNA-1, miRNA 92a, miRNA-133a, and miRNA-133b identified as the most significant diagnostic markers.
- Serum-based miRNA signatures related to BC were successfully validated and derived. |
Sochor et al., 2014 [43] |
- EBC Patients sera = 63 and HC= 21.
- Timing of sera collection: one-day prior operation (time point I), 14–28 days after operation before any non-surgical treatment (time point II) and 14–28 days after first treatment modality (time point III). Upon clinical relapse the patients (N = 3) donated another serum sample (time point IV). |
- RNA extraction: miRNeasy® Mini Kit.
- RT was done by using High Capacity cDNA Reverse Transcription Kit supplemented with miRNA-specific primers. |
- Surgery had drastically effect on three serum oncomiR (miRN A-155, miRNA-181b, and miRNA-24) levels, whilst adjuvant therapy mainly reduced all of four oncomiR (miRNA-155, miRNA-181b, miRNA-24 & miRNA-19a). |
Erbes et al., 2015 [44] |
- MSU gathered in a case–control cohort of:
1) 24 untreated patients, 2) newly diagnosed with primary BC, 3) 24 HCs.
- Serum samples of four consecutive patients and HCs. |
- RNA isolation:
Urine: Norgen’s Urine microRNA Purification Kit, Serum: Norgen’s kit.
- Pre-amplification: augment of miRNA-cDNA quantity was attained by application of Megaplex™ PreAmp Primers, Human Pool A.
- MiRNA expression levels: quantitative real-time-PCR applying TaqMan® MicroRNA Assays. |
- MiRNA-21, miRNA-125b, miRNA-451 and miRNA-155altered and regulated in BC patients in comparison with HCs.
- MiRNA 155 levels were higher, while miR-21, miRNA-125b and miR-451 levels were low in BC patients compared to HC.
- May be urinary miRNAs play a potential role as non-invasive innovative urine-based biomarkers for BC detection. |
Shimomura et al., 2016 [45] |
- Sera samples:
BC= 1280, NCC= 2836, other types of cancer= 451, non-breast benign diseases= 63.
- The samples divided into: 1) Training cohort (NCCs, other cancers and BC)
2) Test cohort (NCCs & BC). |
- Total RNA: extracted from a 300 µL serum sample using 3D Gene RNA extraction reagent from a liquid sample kit.
- Comprehensive miRNA expression analysis: done by using a 3D-Gene miRNA Labeling kit and a 3D-Gene Human miRNA Oligo Chip.
- miRNA to be present:
The corresponding microarray signal was more than the (mean + 2 × standard deviation) signal of the negative controls.
- Quantitative RT-PCR assay: TaqMan microRNA assay |
- A collection of miRNA-1246, miRNA-1307-3p, miRNA-4634, miRNA-6861-5p, and miRNA-6875-5p assessed from serum can be used to survey EBC and to differentiate BC from pancreas ⁄ biliary tract ⁄ prostate benign diseases or other cancers. |
Tsai et al., 2018 [46] |
- First stage:
10 samples from 7 young BC patients (tumor: n = 6, normal tissue: n = 4) have been analyzed by an Agilent microRNA 470 probe microarray.
- Second stage:
85 candidate miRNAs have been validated in 145 BC in 4 subgroups:
1- Very young BC (n=15, ≤35 y)
2- Young BC (n=37, 36-40 y)
3- Premenopausal BC (n= 47, 41-50 y)
4- Postmenopausal BC (n=46, >50 y) |
- Total RNAs have been extracted from tissue by using TRIzol.
- 85 miRNAs were selected for qPCR by Agilent microRNA 470 probe microarray through two ways:
1- MiRNAs that the fold-change and the t-test p value were > 1.5 and < 0.05 between tumor and normal breast tissues (44 miRNAs).
2- MiRNAs that have been reported
in the literature to be related to BC (41 miRNAs). |
- 6 miRNAs had significantly differential expression, 2 out of 6 have been down-regulated, including miRNA-145 and miRNA-335, whilst 4 out of 6 have been up-regulated which consist of miRNA-21, miRNA-200a, miRNA-200c and miRNA-141.
- As well as, the below detections have been seen separately in each subgroup:
1- In very young BC 3 miRNAs (miRNA-96, miRNA-182, and miRNA-183) and 8 miRNAs (miRNA-10a, miRNA-10b, miRNA-125b, miRNA-127-3p, miRNA-130a, miRNA-143, miRNA-19, and miRNA-320) have been down- and up-regulated, respectively.
2- For young BC, upregulation of 3 miRNAs (miRNA-30b, miRNA-30d, and miRNA-149) and downregulation of 3 miRNAs (miRNA-487b, miRNA-548c-5p, and miRNA-181d) have been observed.
3- Only miRNA-206 has been down-regulated in premenopausal BC.
4- No alteration has been observed in postmenopausal BC. |
MeDIA: Methylated DNA isolation assay; MBD2bt: Methyl DNA binding domain; MeDIP: Methyl cytosine DNA Immunoprecipitation; BC: Breast cancer; CGI: CPG island; ERα-: Estrogen receptor α-negative; MSP: Methylation-specific PCR; cfDNA: Cell-free DNA; HER2: Tyrosine kinase-type cell surface receptor; TNBC: Triple-negative breast cancer; HRD: Homologous recombination deficiency; FFPE: Formalin-fixed, paraffin-embedded sections; FOXF2: Fork-head box F2; BCFR: Breast cancer family registry; SNP: Single-nucleotide polymorphisms; TCGA: The cancer genome atlas; ROC: Receiver operating characteristic; TLDA: TaqMan low-density arrays; FDR: False discovery rate; RT: Reverse transcription; NATs: Normal adjacent tissues; HC: Healthy control; LNA RT-PCR: Locked nucleic acid real-time PCR; EBC: Early-stage breast cancer; MSU: Midstream specimen of urine; NCC: Non-cancer control; qPCR: Quantitative real-time PCR analysis
Table 2. miRNA and their role in various cancer
|
|
miRNA type |
Cancer type |
Comment |
OncomiRs |
miR17-92 [63]
(miR-17, miR-18a, miR-19a, miR-19b, miR-20a, and miR-92a) |
B-cell lymphoma, colon and gastric cancer. |
Acts as multi-targeted regulators of T-ALL tumor suppressors including PTEN, BIM, NF1, FBXW7, IKZF1 and PHF6 |
miR-21 [64] |
Breast, colon, lung cancer, and glioblastoma, NSCLC. |
Increases growth, promote migration & invasion, and enhance chemo- or radio- resistance, metastasis. |
miR-106 [65] |
Gastric and colorectal cancer |
Proliferation of tumor cells. |
miR-10b [66] |
Breast and Esophageal Cancer |
Cell motility and invasiveness. |
miR-191 [67] |
Human colorectal and BC |
Promoting cell proliferation and migration (by inducing TGF-β2 expression). |
Tumor suppressor miRNAs |
let-7 [68] |
Lung cancer and Burkitt lymphoma |
Engaged in the proliferation, apoptosis and invasion of cancer cells; induced by EphrinA1 and inhibits RAS. |
miR-15a, miR16-1 [69] |
CLL, prostate cancer, and mesothelioma |
MiR-15a: Inhibits growth of MPM cells.
MiR16-1: Downregulates cell cycle and anti-apoptotic genes CCND1 and BCL2. |
miR-29 [70] |
Lung and breast cancer |
MiR-29a reduces the invasive phenotype of cancer (miR-29a expression in the invasive variant was less than 45% compared to non-invasive cell line). |
miR-34a [71] |
Prostate cancer, HCC, and mesothelioma. |
Engaged in the regulation of different cancer processes, such as apoptosis, proliferation, invasion, migration, and metastasis. |
miR-126 [72] |
Lung and breast cancer |
Perhaps acts as a tumor suppressor via prohibition of cell invasion by downregulating ADAM9 (metalloprotease 9). |
Both OncomiRs and Tumor suppressor miRNAs |
miR-24 [73, 74] |
NSCLC (O)
LSCC (TS) |
Acts as oncomir through regulation of its functional target NAIF1.
Expression of miR-24 in Hep2 cells triggered cell morphology alterations and prohibited cell invasion and proliferation ability by targeting S100A8 at the translational level. So through under-expression of S100A8 in LSCC, miR-24 act as TS. |
miR-125 [75]
(hsa-miR-125a, hsa-miR-125b-1 & hsa-miR-125-2) |
Pancreatic, prostate and oligodendro glial cancer (O)
Melanoma, osteosarcoma, ovarian, bladder, and breast cancer (TS) |
Members of the family have main roles in cell proliferation, differentiation, and apoptosis by targeting matrix-metalloprotease, transcription factors, and growth factors. |
miR-155 [76] |
Lymphoma and BC (O)
Melanoma, ovarian and gastric cancer (TS) |
Up-regulation of miR-155 can restrain melanoma growth through down regulation of SKI protein, so improve proliferation and apoptosis.
Also, upregulation of miRNA 155 in SGC-7901 and MKN-45 gastric cancer cells suppressed cell adhesion, invasion, and migration. |
miR-221/222 [77] |
Glioblastoma, HCC, BC, and NSCLC (O)
OTSCC and Erythrocytes (TS) |
MiR-221/222 acts as oncomir through regulation of its functional target P57 and DDIT4 (HCC), Puma (glioblastoma), ER-α and FOXO3 (Breast), PTEN (NSCLC).
MiR-221/222 acts as TS through regulation of its functional target SODI MMP1 (OTSCC), c-Kit (Erythrocytes). |
SCLC: Small cell lung cancer; T-ALL: T-cell acute lymphoblastic leukemia; NSCLC: Non-small cell lung cancer; CLL: Chronic lymphocytic leukemia; MPM: Malignant pleural mesothelioma; HCC: Hepatocellular carcinoma; EMT: Epithelial to mesenchymal transition; LSCC: Laryngeal squamous cell carcinoma; NAIF1: Nuclear apoptosis-inducing factor 1; OTSCC: Tongue squamous cell carcinoma; O: OncomiR; TS: Tumor suppressor miRNA.
Table 3. Summary of nutrigenomics and BC studies
References |
Nutrient components |
Major finding |
Roystone et al. (2018) [104] |
5.0 µM SFN, 1.0 µM WA or both. |
- In MCF-7 cells: SFN or WA reduced transition into the G2/M phase, whereas the combination of them arrested cells at the G1 phase.
- In MDA-MB-231 cells: SFN+WA showed a decline in transition into S phase, and SFN consider to be more effective than WA.
- CCND1 and CDK4 (promote transition from G1 to S phase) expression reduced with the SFN and WA alone and also pRB (G1 checkpoint) decreased, while the combination of SFN+WN was not most effective in both cell lines BC cells.
- Up-regulation of E2F CDK4 (promote transition from G1 to S phase) in both cell lines was most effective by SFN+WA.
- WA increased HAT activity in both cell lines but it was significant in MDA-MB-231 cells, in comparison with SFA+WA and SFA.
- SFA+WA significantly increased global methylation in both MCF-7 and MDA-MB-231 cells.
- P21expression increased in MDA-MB-231 cells, and P-53 reduced by SFN+WA. Whereas, it was observed mild go up in P21 and P53 in MCF-7 cells by SFN or WA and both of them.
- SFN or WA alone, or in combination with WA mainly augmented the surfaces of H3K4Me3 at the p21 promoter, showing that these nutrient compounds positively modulate the epigenome at the promoter region. |
Zhu et al. (2012) [105] |
- supplementing with capsule twice daily for 12 weeks containing 1 of 3 preparations:
1-placebo, 2-P. cuspidatum with 5 mg trans-resveratrol, 3-P. cuspidatum with 50 mg of trans resveratrol |
- RASSF-1α (BC related gene) methylation declined with increasing levels of serum trans-resveratrol. |
Qin et al. (2014) [106] |
- Rats were randomly divided in 4 groups:
1- Control group,
2- Low dose resveratrol (lo; 5 mg/kg/day),
3- High dose resveratrol (hi; 25 mg/kg/day),
4- Resveratrol in a Harlan Teklad prepared diet, or 0.1 mg/kg/wk 5-aza-2 deoxycytidine by intraperitoneal injection, |
- Hi resveratrol and Aza provoked a differential impact on DNMT3b expression in tumor in comparison with normal tissue. DNMT3b reduced in tumor but up-surged in normal tissue.
- Lo but not hi resveratrol augmented miR10a and miR10b expression in tumor tissue.
- Hi dose augmented miRNA-21, -129, -204, and -489 and reduced the same miRs in normal tissue.
- It was observed contradict association between DNMT3b and miRNA-129, -204, and -489 in normal and/or tumor tissue. |
D. Adams et al. (2018) [107] |
- |
- Four miRNAs, including miRNA-22-3p, miRNA-122-5p, miRNA-126-3p, miRNA-150-5p have positive correlation with BMI, whilst miRNA-191-5p, miRNA-17-5p, miRNA-103a-3p, miRNA-93-5p have negative association with BMI in HOPE trial.
- Expression of miRNA-106b-5p, let-7b-5p, and miRNA-92a-3p decreased, whereas miRNA-27a-3p, miRNA-191-5p, miRNA-24-3p increased at the end of the six-month intervention by arm in the LEAN trial. |
Pietruszewska et al. (2015) (108) |
- MCF-7 and MDA-MB-23 cells cultured for 96 h with SFN at different concentrations. |
- In MCF-7 cells at 10 and 22 μM concentrations of SFN: PTEN and RARbeta2 promoter methylation decreased by 25 and 12 %, respectively, whilst PTEN mRNA upregulated by 32% in 10 μM and 82% in 22 μM and RARbeta2 expression increased 55–60%.
- In MDA-MB-231 cells at 10 and 46 μM concentrations of SFN: decreased in PTEN promoter methylation by 22 and 31%, and PTEN mRNA expression increased by 72 and 101%, respectively.
- In MCF-7 cells, p21 expression increased by 2.5- and 1.8-fold, and p53 mRNA levels augmented by 66 and 38% at 10 and 22 μM SFN concentrations, respectively.
- In MDA-MB-231 cells which express mutant p53, SFN at 10 and 46 μM concentrations enhanced p21 expression by 3-fold and 55%, respectively.
- Although regulators of DNMT1, p21 and p53 over-expressed, there was no effect on DNMT1 suppression. |
Peng et al. (2017) [109] |
20 mg/ kg/day of 3,6-DHF |
- 3,6-DHF could reverse the global down-regulation of miR-34a in BC by regulating the miR-34a promoter methylation.
- 3,6-DHF up-lifted TET1 level in MDA-MB-231 cells, and had no influence on TET2 and TET3.
- 3,6-DHF prohibited DNMT1 activity, modified the disequilibrium of DNA methylation and demethylation status. |
Kumar et al. (2017) [110] |
- MCF-7 cells treated with curcumin at 0, 1, 3, 5, 10, 20, 30, 50, and 100 µM for different time periods, including 24, 48, and 72h. |
- Treated MCF-7 cells with 10 µM of curcumin for 72h reversed the GSTP1 promoter hyper-methylation and re-expressed GSTP1 protein. |
Pietruszewska et al. (2013) [111] |
- ER-positive MCF-7 and invasive ER-negative MDAMB-231 cells, treated with 1 mg/l in control and 4 mg/l and 8 mg/l in experiment groups. |
- In high dose administration of folic acid, down-regulation of tumor suppressor genes, including PTEN, APC and RARbeta2 and increasing DNA methylation in their promoters occurred in MCF-7 cells.
- Induced DNMT1 expression in MCF-7 cells in highest dose. |
SFN: Sulforaphane; WA: Withaferin A; CCND1: Cell cyclin D1; CDK4: Cyclin D kinase 4; pRB: phosphorylated retinoblastoma protein; HAT: Histone acetyltransferase activity; H3K4Me3: Trimethylated lysine 4 of histone; HOPE: Hormones and physical exercise; LEAN: Lifestyle, exercise, and nutrition; SFN: sulforaphane; 3,6-DHF: 3,6-dihydroxyflavone; TET: Ten-eleven translocation; GSTP1: Glutathione S-transferase pi 1.




