In 2020, a very powerful web-server predictor has been established for identifying the subcellular localization of human proteins based on the sequence information alone [1], in which a same protein may occur or move between two or more location sites and hence needs to be marked with the multi-label approach [2].
The web-server predictor is called “pLoc_Deep-mVirus”, where “Deep” means the web-server has been further improved by the “Deep Learning” technique [3-6], and “m” means the capacity able to deal with the multi-label systems. To learn how the web-server is working, please do the following.
Step 1. Click the link at http://www.jci-bioinfo.cn/pLoc_Deep-mVirus/, the top page of the pLoc_bal-mVirus web-server will appear on your computer screen, as shown in Figure 1. Click on the Read Me button to see a brief introduction about the predictor.
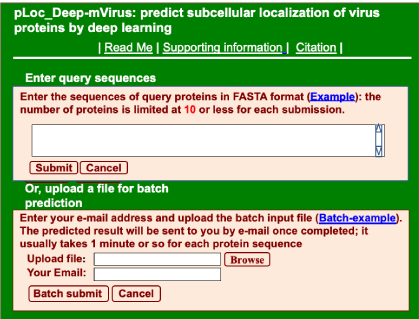
Figure 1. The pLoc_bal-mVirus web-server
Step 2. Either type or copy/paste the sequences of query human proteins into the input box at the center of Figure 1. The input sequence should be in the FASTA format. For the examples of sequences in FASTA format, click the Example button right above the input box.
Step 3. Click on the Submit button to see the predicted result. For instance, if you use the four protein sequences in the Example window as the input, after 10 seconds or so, you will see a new screen (Figure 2) occurring. On its upper part are listed the names of the subcellular locations numbered from (1) to (6) covered by the current predictor. On its lower part are the predicted results: the query protein P01115 of example-1 corresponds to “2,” meaning it belongs to “Host cell membrane” only; the query protein P03495 of example-2 corresponds to “4, 5” meaning it belongs to “Host cytoplasm” and “Host nucleus”; the query protein P89873 of example-3 corresponds to “4, 5, 6”, meaning it belongs to “Host cytoplasm”, “Host nucleus”, and “Secreted”. All these results are perfectly consistent with experimental observations.
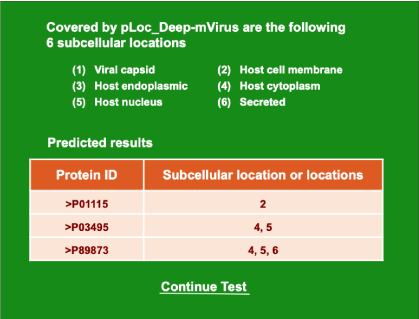
Figure 2. Continue test
Step 4. As shown on the lower panel of Figure 2, you may also choose the batch prediction by entering your e-mail address and your desired batch input file (in FASTA format of course) via the Browse button. To see the sample of batch input file, click on the button Batch-example. After clicking the button Batch-submit, you will see “Your batch job is under computation; once the results are available, you will be notified by e-mail.”
Step 5. Click on the Citation button to find the papers that have played the key role in developing the current predictor of pLoc_Deep-mVirus.
Step 6. Click the Supporting Information button to download the Supporting Informations mentioned in this paper.
- Shao YT, Cheng X, Chou KC (2020) pLoc_Deep-mVirus: a CNN model for predicting subcellular localization of virus proteins by deep learning Molecular Therapy - Nucleic Acids (MTNA), in press.
- Chou KC (2019) Advance in predicting subcellular localization of multi-label proteins and its implication for developing multi-target drugs. Curr Med Chem 26: 4918-4943. [Crossref]
- Maxwell A, Li R, Yang B, Weng H, Ou A, et al. (2017) Deep learning architectures for multi-label classification of intelligent health risk prediction. BMC Bioinformatics 18: 523. [Crossref]
- Khan S, Khan M, Iqbal N, Hussain T, Khan SA, et al. (2019) A Two-Level Computation Model Based on Deep Learning Algorithm for Identification of piRNA and Their Functions via Chou's 5-Steps Rule. Human Genetics 26: 795-809.
- Nazari I, Tahir M, Tayari H, Chong KT, iN6-Methyl (5-step): Identifying RNA N6-methyladenosine sites using deep learning mode via Chou's 5-step rules and Chou's general PseKNC. Chemometrics and Intelligent Laboratory Systems 193: 103811.
- Khan ZU, Ali F, Khan IA, Hussain Y, Pi D (2019) iRSpot-SPI: Deep learning-based recombination spots prediction by incorporating secondary sequence information coupled with physio-chemical properties via Chou's 5-step rule and pseudo components. Chemometrics and Intelligent Laboratory Systems 189: 169-180.


