Food products of animal origin such as fresh meat are easily contaminated by microorganisms if handling, processing and storage conditions are not fully respected. The present study aimed first to evaluate the bacterial load and microbial contamination rates of ground raw beef to identify the main pathogenic flora that dominate and second, to determine the resistance patterns and extended-spectrum beta-lactamase (ESBL) of isolated Gram-negative strains against certain families of antibiotics. Therefore, 39 samples have been collected from 5 butcher shops located in Constantine province in the North-East of Algeria. The samples were analysed for total bacterial count, presence of total and faecal coliforms, Staphylococci and Salmonella. Furthermore, 23 antibiotics were tested using the diffusion method on Mueller-Hinton agar, towards 22 strains isolates. Bacterial analyses showed a high contamination by total aerobic bacteria, total and faecal coliforms. Escherichia coli, Citrobacter spp., Enterobacter spp., Hafnia alvei, Salmonella pullorum and Staphylococcus spp (except Staphylococcus aureus) were further revealed in some samples. The results of the antibiogram test exhibit multi-resistance to more than eight antibiotics with varied effects. From the whole tested strains isolates, the fully susceptibility effect was for spectinomycin (SPT). This study reveals that the analysed minced meat was found to be highly contaminated with antibiotic resistant bacteria. This study allows concluding that appropriate use of antibiotics in compliance with good hygiene practices is essential to reduce the antibiotic resistance identified in this preliminary study.
minced meat, contamination, bacteria, antibiotics, safety, Algeria
Red meat constitutes an important part in the human diet throughout human evolution, considered as the major food protein source [1]. Muscle tissues of carcasses from healthy animals are essentially free of bacteria until they are exposed during skinning, where the transfer of bacteria from the hide and the environment to the meat is inevitable. Thus, fresh meat is easily damaged and highly contaminated [2]. Human pathogen contamination of raw meat products is caused by a wide array of pre-harvest, harvest, and post-harvest processes [3]. Although thorough cooking kills pathogens, cooked meat may become re-contaminated by food handlers during processing or from the environment. Indeed, contamination of raw meat with pathogens, such as Salmonella spp., Campylobacter spp., Listeria monocytogenes, Staphylococcus aureus or Escherichia coli, currently considered, as potential sources of food-borne infections or food poisoning could result in adverse effects on human health [1]. Food-borne diseases represent an important public health problem, significantly affecting the health of the population with major economic consequences. In this respect, the contamination of food with zoonotic bacteria is just one among many issues [4]. Due to potential food safety concerns associated with meat products, the food industry is strongly ask to assess the potential risk mitigation strategies that would reduce pathogen populations on raw meat.
On another hand, the resistance to antibiotics in food-borne pathogens may create problems for disease or illness treatment. Indeed, the extensive and intensive use of antibiotics in the veterinary medicine for treatment of sick animals or for prophylactic purposes contributed to the emergence of resistant bacteria, that can be transmitted in the food chain to human [5].
In this context, this preliminary study aimed to assess the microbial contamination of fresh minced beef in the Northeast of Algeria, the case of butchers from Constantine, and to determine the prevalence of antibiotic-resistant bacteria, and extended-spectrum beta-lactamase (ESBL).
Sample collection
Thirty nine samples of minced raw beef were randomly collected in sterile bags from five butcher shops located in Constantine province, Northeast of Algeria. They were iced and transported to the laboratory for immediate analysis.
Isolation of microorganisms
Under aseptic conditions, 25 g of each sample were weighed and homogenized in a stomacher blender (150 rpm/min for 2 min) with 225 mL of 0.1% peptone water (pH 7.0). The procedure described by Boudechicha et al. [6] was followed to count total aerobic bacteria, total and faecal coliforms, Staphylococcus aureus in each sample (duplicates). Furthermore, Salmonella spp. was searched.
The culture methods for the detection of different organisms were based on the following international standards:
- EN ISO 4833-2003 for total Mesophilic Aerobic Counts
- ISO 21528-2- 2004 for Enterobacteriae
- NF V 08-060-1996 for Thermotolerant coliforms counts.
- ISO 6888-1-1999 for Coagulase positive Staphylococcus aureus counts.
- EN ISO 6579-2007 for the detection of Salmonella species.
Five colonies per sample were randomly selected and identified using API 20 E and API Staph (Biomérieux. Marcy l’Etoile, France).
Antibiotics susceptibility test
All the isolated strains were tested for their susceptibility against 23 antibiotics, using the Kirby-Bauer antimicrobial disk diffusion procedure on Muller-Hinton agar. The results were interpreted according to the Committee of the French Society of Microbiology (CA-SFM 2019) [7]. The antimicrobial disks (Biomérieux, Marcy L’étoile, France) tested, contained the following antibiotics: Amoxicillin (AML) (25 µg), Amoxicillin-clavulanic (AMC) (20/10 µg), Ampicillin (AMP) (25 µg), Carbenicillin (CAR) (100 µg), Oxacillin (OXA) (5 µg), Penicillin (PEN) (6 µg), Cloxacillin (CLX) (10 µg), Cephalothin (CEF) (30 µg), Cefoperazon (CFP) (75 µg), Cefoxitin (FOX) (30 µg), Amikacin (AMK) (30 µg), Streptomycin (STR) (10 µg), Gentamicin (GEN) (10 µg), Kanamycin(KAN) (30 µg), Neomycin (NEO) (30 µg), Apramycin (APR) (15 µg), Tobramycin (TOB) (10 µg), Spectinomycin (SPT) (10 µg), Chloranfenicol (CHL) (30 µg), Fosfomicyn (FOF) (50 µg), Tetracyclin (TE) (30 µg), Sulfamethoxazole-trimethoprim (SXT) (25 µg) and Mupirocin (MUP) (5 µg).
Determination of B-lactamase enzyme production
The double-disk diffusion synergy test (DDST) was used according to Farrag et al. [8] and Iqbal et al. [9] as screening methods to detect the production of extended-spectrum B-lactamase (ESBL) and Ampc-B-lactamases by selected strains. The test was performed using disks of FOX (30 μg), CFP (75 μg), CEF (30 μg) and AMC (30 μg). At the centre of the disc, AMC was placed and these discs were placed at a distance of 1.5 cm. The development of the zone of inhibition towards the Clavulanate disc at 37°C after 24h incubation was indicative of a potential ESBL positive organism.
Statistical analysis
The statistical software XLSTAT 2017.19.4 (AddinSoft, Paris, France) was used to perform one way ANOVA tests in order to compare the contamination levels (with the different bacterial groups) to compare the samples from the butchers sampled at different months. For comparisons, the Tukey's test at a significance level of P < 0.05 was used. All the microbiological analyses were carried out in duplicate and the results were expressed as log10 cfu/g.
Distribution and prevalence of the main identified microorganisms in the meat samples
The distribution of bacterial species per butcher is summarized in Table 1. It showed high contamination levels of the marketed minced meat with total aerobic bacteria and total and faecal coliforms. The biochemical identification confirmed the presence of E. coli, Salmonella spp, Enterobacter cloacae, Citrobacter spp, Hafnia alvei and Staphylococcus spp. However, none of the Staphylococcus aureus strain was isolated.
Table 1. Summary of the bacterial species present per butcher shop
Butchers |
Bacterial species |
Butcher 1 (n=7) |
Staphylococcus xylosus
Citrobacter brakii
Citrobacter frendii
E. coli |
Butcher 2 (n=7) |
Citrobacter brakii
Staphylococcus xylosus
E. coli |
Butcher 3 (n=9) |
Enterobacter cloacae
Citrobacter brakii
Staphylococcus xylosus
Staphylococcus hominis
E. coli |
Butcher 4 (n=8) |
E. coli
Salmonella pullorum
Citrobacter brakii
Citrobacter frendii |
Butcher 5 (n=8) |
Hafnia alvei
E. coli |
The prevalence of contamination of minced meat is around 50% according to the JORA (Official Journal of the People's Democratic Republic of Algerian, 2017). Staphylococcus spp. were the dominant bacteria isolated in this study as they were identified in 87.5% of samples, followed by total coliforms (75%) and faecal coliforms (50%), respectively. The mean contamination rates (Log cfu/g) per butcher shop and per sampling month are shown in Table 2 and Table 3, respectively.
Table 2. Bacterial groups’ rates (Log CFU/g) within the minced raw beef samples collected from five different butchers
Sampled butcher |
Total aerobic bacteria |
Total Coliforms |
Faecal Coliforms |
Staphylococcus spp 1 |
Butcher 1 (n=7) |
5.68±1.1 |
5.46±1.4 |
5.16±1.0 |
3.47a±1.4 |
Butcher 2 (n=7) |
5.77±0.8 |
5.46±1.3 |
5.16±0.8 |
2.73b±1.3 |
Butcher 3 (n=9) |
5.42±0.6 |
4.07±1.9 |
3.73±1.7 |
3.48a±1.9 |
Butcher 4 (n=8) |
5.48±1.2 |
5.41±0.6 |
5.14±1.0 |
2.18b±1.6 |
Butcher 5 (n=8) |
5.30±0.4 |
3.70±2.6 |
2.44±1.6 |
2.38b±2.1 |
Significance 2 |
ns |
ns |
ns |
* |
1Least square means in the same column not followed by a common letter differ significantly at P<0.05.
2Significance levels: ns: not significant (P > 0.1); *P<0.05 |
Table 3. Bacterial groups’ rates (Log CFU/g) within the minced raw beef samples collected at four different months of the same year
Sampling month |
Total aerobic bacteria |
Total Coliforms |
Faecal Coliforms |
Staphylococcus spp |
November (n=3) |
5.03±0.4 |
5.43±1.2 |
3.06±1.7 |
3.09±0.9 |
December (n=17) |
5.99±1.0 |
5.89±1.2 |
5.06±1.2 |
3.19±1.1 |
March (n=10) |
5.43±1.6 |
3.58±1.4 |
4.56±1.1 |
2.37±1.7 |
April (n=10) |
4.69±2.3 |
5.04±0.4 |
5.13±0.8 |
2.69±1.4 |
Significance1 |
ns |
ns |
ns |
ns |
1Significance levels: ns: not significant (P > 0.1) |
The microbiological analyses of the minced meat samples provided us with information on the hygienic quality commercialised product. Indeed, the use of qualitative and quantitative methods, has allowed the detection of some pathogens and the determination of high contamination rates with other indicator microorganisms such as Enterobacteriaceae, coliforms and Enterococci.
The contamination rates reported in this study are lower than those obtained by Bouzid et al. [10] who found 96.6% of minced raw beef (n = 60) collected in Western Algeria to be contaminated with total and faecal coliforms, Salmonella and Staphylococcus aureus. The prevalence of this study was further different from that by Oumokhtar et al. [11] in Morocco, reporting a contamination rate of 80% with total and faecal coliforms including E. coli, Salmonella, Shigella and Staphylococcus aureus.
Citrobacter spp. was isolated in 15% of samples, E. coli in 32.5%, Hafnia alvei, Enterobacter spp. and Salmonella pullorum in 2.5% of the samples. These findings are in contrast to those reported by Tassew et al. [12] who analysed 165 samples of minced beef in Ethiopia and identified E. coli in 26.6% of them, Citrobacter spp in 9%, Enterobacter spp. In 1.2%, Salmonella spp in 1.2% and Staphylococcus aureus in 12.1%. The results of this study are also different from those reported by El-Gendy et al. [13] (Alexandria, Egypt) who noticed that the prevalence of E. coli was 44%, Enterobacter aerogenes 12%, Enterobacter intermedium 4%, Enterobacter gergoviae 4%, Citrobacter amalonaticus 4%, Citrobacter diversus 4%, Citrobacter freundii 4%, Serratia marcescens 8%, Serratia ficaria 8%, Serratia fonticola 12%, Serratia liquefaciens 4 %, Serratia rubidaea 8%, Edwardsiella ictalori 8%, Edwardsiella hoshinae 12%, Providencia alcalifciens 4%, Klebsiella pneumonia 4% and Proteus mirabilis 16% in a total of 100 samples. Another recent study by Yusuf et al. [14] found different prevalence for E. coli (28.6%), Enterobacter spp. (24.5%), Pseudomonas aeruginosa (14.3%), Salmonella spp. (6.1%) and Staphylococcus aureus (16.3%).
Table 1 summarises that the distribution of the isolated bacterial strains is different from one butcher shop to another with the lowest contamination rate observed for butcher shop number 5. Surprisingly Staphylococcus spp. was isolated in only three butcher shops (number 1, 2 and 3). Indeed, the statistical study given in Table 2 corroborates with this observation and demonstrates that no significant difference (P > 0.05) in contamination rates exists between the five butchers with all the tested bacteria except Staphylococcus spp (P < 0.05).
According to Joffin and Joffin [15], the detection and the enumeration of Staphylococcus spp. is used to evaluate the bacterial risk to consumers, as it is the main bacteria that can produce an enterotoxin responsible for food poisoning.
Investigations done by Zerabruk et al. [2] reported a heavy bioburden of Staphylococci. The high contamination by Staphylococci could be associated with improper personal hygiene of untrained employees and a cross contamination from skin and utensils.
From Table 2, there is no significant difference (P > 0.05) among the period of sampling (4 months). It seems that there is a tendency of the lowest microbial contamination levels during March and April. These results indicate that the contamination of the samples was influenced by neither the change of season, nor the increase in temperature.
In this study, bacterial contamination with total aerobic bacteria per butcher and per sampling month were under the standard level fixed by the Algerian guidelines [16]. According to Lasta et al. [17], the total plate count informs about the overall degree of contamination of meat.
Alarmingly, the results of this study showed the presence of micro-organisms considered pathogenic to human such as Salmonella, E. coli and Staphylococcus aureus [10]. According to Zweifel et al. [18], the determination of Enterobacteriaceae is an essential element in assessing the quality of marketed meat. Our samples are more contaminated with Enterobacteriae (by butchers or by sampling month) than the level permitted by the Algerian regulations [16]. Since members of Enterobacteriaceae are safety indicators, their high counts may be associated with the possible presence of potential pathogens [19].
The high rate of total coliforms in minced meat may be due to improper cleaning, contaminated materials and poor storage. While the high presence of faecal coliform would reflect the poor conditions during the slaughter process, the presence of total coliforms indicates recent faecal contamination [10]. Also, the presence of the marker bacterial groups such as coliforms andE. coliin the processed products demonstrates a possible contamination during the process related to poor manufacturing practices and inadequate factory hygiene standards [20].
The presence of Salmonella pullorum in minced meat samples is probably due to a cross-contamination from poultry meat [12,14,21]. In fact, cross-contamination during food preparation has been identified as an important factor associated with food-borne [22]. According to Sousa [21], food-borne caused by Salmonella, E. coli and S. aureus is a major public health problem worldwide. These pathogens are transmitted mainly through consumption of contaminated food. The presence of these microorganisms in foods of animal origin and raw meat products has relevant public health implications [23]. Salmonellais the leading cause of human food-borne infections and poultry meat is one of the main vehicles [14]. The main reason of contamination in this study would be due to the slaughtering process in abattoirs which is usually realized manually and in very poor hygienic conditions.
According to Regab et al. [24], fresh minced meat tends to have a short life due to the aspect of the raw ingredients that are re-contaminated through the grinding/handling process. Mincing and grinding of meat at the retail location can introduce more spoilage microorganisms if proper equipment hygiene and handling measures are not respected.
Antibiotics susceptibility test and determination of B-lactamase enzyme production
The results of this study revealed multi-resistance drugs (Figure 1). Indeed, the resistance is 100% to Ampicillin (AMP), Oxacillin (OXA), Cloxacillin (CLX), Mupirocin (MUP); 98.72% to Apramycin (APR), 95.45% to Penicillin (PEN), 90.90% to Amoxicillin (AML), 86.36% to Tobramycin (TOB) and Neomycin (NEO); 68.18% to Cephalothin (CEF), 50% to Amoxicillin-clavulanic (AMC) and Gentamicin (GEN); 45.45% to Cefoxitin (FOX) and Streptomycin (STR), 27.57% to Tetracyclin (TE), 22.72% to Carbenicillin (CAR), 13.63% to Chloranfenicol (CHL) and Kanamycin (KAN); 10.90% to Sulfamethoxazole-trimethoprim (SXT),4.54% to Fosfomicyne (FOF), Amikacin (AMK) and Cefoperazon (CFP). All tested strains were susceptible to spectinomycin (SPT) (Figure 1).
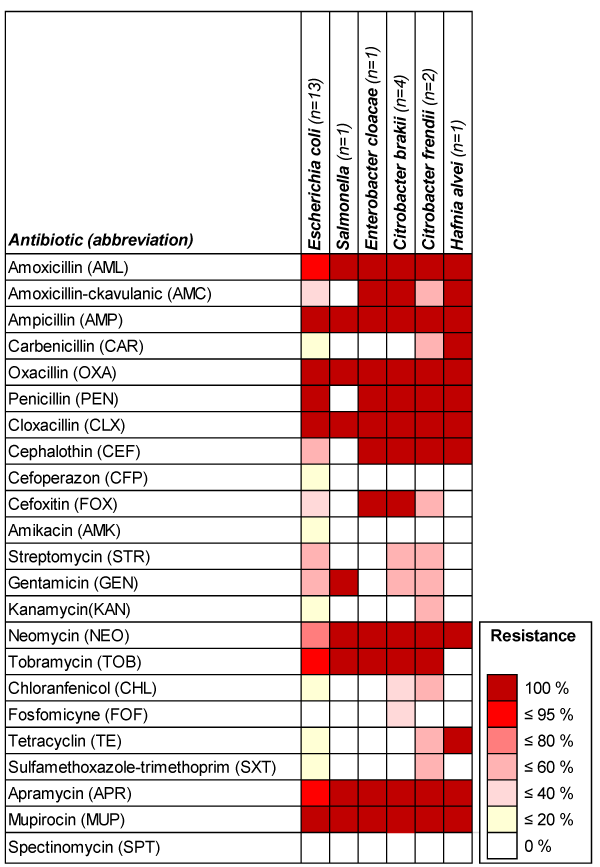
Figure 1. Antimicrobial resistance of the isolates of bacterial species against different antibiotics
The results obtained in this study are different from those by Hassan et al., [25] who observed in the strains isolated from raw meat (n = 250) in Pakistan, 72% resistance to Ampicillin, 75% to Amoxicillin, 70% to Cefaclor and 62% to Novobiocin. A total of 50% of the isolates were resistant against Roxithromycin, while only 33% were resistant against Cephalexin.
The results of this study allowed to see that the antibiotic resistance Enterobacteriaceae is high. Indeed, all the bacterial species showed a resistance to over 8 antibiotics (Table 4). Thus, the isolated strains have a different resistance profiles comparing to those reported by Hassan et al. [25] who observed a resistance to more than 10 antibiotics in samples of raw meat which, included 35% of E. coli, 8% of Klebsiella, 15% of Enterobacter spp. and 7% of Staphylococcus aureus. Antibiotic resistance is a worldwide health problem in human and veterinary medicine [26]. According to the World Health Organization (WHO), this resistance is due to the ability of the bacterial population to survive to the effect of an inhibitory concentration of antimicrobial agents [27-29].
Table 4. Bacterial species resistance phenotypes depending on different antibiotics families
The results in Table 4 highlighted the multidrug resistance (MDR) of all species tested. However, the strains studied are sensitive to SH and PSC. This MDR is more common than the resistance to one antibiotic and has become one of the biggest challenges in clinical therapy [30,31]. Recent studies showed that the antibiotic-resistant bacteria are not only present in farm animals, but also in the environment [32] and even in air [4,33]. According to Doyle [34], antimicrobial resistance, including MDR, is an increasing problem globally. MDR bacteria are frequently detected in humans and animals from both more-and less-developed countries and pose a serious concern for human health. Infections caused by MDR microbes may increase morbidity and mortality and require use of expensive drugs and prolonged hospitalization. Human may be exposed to MDR pathogens through exposure to environments at health-care facilities and farms, livestock and companion animals, human food, and exposure to other individuals carrying MDR microbes. Indeed food-associated MDR may be an emerging problem [35,36].
The resistance phenotypes, by family of antibiotics are summarized in Table 4. The families of antibiotics resistance phenotypes are different from one bacterial species to another and from one strain to another. The studied bacterial species have different phenotypes of resistance to beta lactams (penicillins and cephalosporins). This resistance may be due either to the decrease in membrane permeability, modification of the target beta lactam antibiotics or to the enzymatic inactivation of beta lactam [37,38]. In the large literature, it has been demonstrated that the production of beta-lactamases is the essential mechanism of resistance to beta lactams in many Enterobacteria. Indeed, the presence of beta-lactamases, enzymes that inactivate this group of antibiotics is always suspected when the isolated gram-negative bacteria are resistant to beta lactam antibiotics. In fact, penicillins, cephalosporins and carbapenems are among the most commonly used and combined treatment of many infectious diseases and the existence of these enzymes play an important role in the selection of effective treatment [39,40]. It was also observed that the bacteria studied showed aminoglycoside resistance phenotypes.
The results of using a double disk diffusion method allowed to see that there is a synergy between CFP and FOX in 16 strains (13 E. coli, 1 Salmonella Pullurum, 1 Citrobacter frendii, and 1 Hafnia alvei), and among the disks containing CFP and AMC in 6 strains (1 E. coli, 4 Citrobacter brakii and 1 Salmonella pullurum) and CFP, CEF adjacent to AMC and FOX disk at distance of 20-25mm from each other in nine strains (8 E. coli and 1 Citrobacter frendii). According to Ruppé et al. [41], the first synergy expresses a plasmid Cephalosporinase ACC-1. The results of the second and the third synergies expressed BLSE in agreement with Carter et al. [42].
In this study, different synergies were observed among the same strains. Thus, the coexistence of different beta lactamases in a single bacterium can cause a diagnostic problem and therapy. These associations lead to a co-selection of resistant genes and pandemic scenario in the hospital and in community [43].
In this study, we were further interested by the determination of beta lactamase depending on phenotype. It has been shown by Courvalin and Philippon [44] that the beta lactamase typing can be established according to the different phenotypes of resistance to beta lactams. The high resistance to penicillin and first and third-generation cephalosporins (C1G, C3G) whose phenotype is the following "AML AMC TIC MEC CTX CF" would cause a beta lactamase extended spectrum (ESBLs). It is worthwhile to note that there are other types of beta-lactamases such as low cephalosporinases "AML AMC CF", high cephalosporinases "TIC AML AMC CF CTX", low penicillinases "AML TIC", and high penicillinases level "AML AMC TIC MEC CF". In our study, Ticarcillin (TIC) has been replaced with Carbenicillin (CAR) (it belongs to the same family of carboxypenicillins) and Ceftazidime (CTX) (C3G) by Cefoperazone (CFP) (C3G), while the Mecillinam has not been tested. These tests allowed us to observe that two strains of E. coli introducing the phenotype “AML AMC CAR CEF” are similar to that of high penicillinases and two other strains of E. coli introducing the phenotype “AML AMP CAR CEF” are similar to that of low penicillinases. Two more strains (1 E. coli, 1 Citrobacter Brakii) introducing the phenotype “AML AMC CEF” are similar to that of low cephalosporinases. Finally, four strains (1 Enterobacter cloacae, 3 Citrobacter brakii) introducing the phenotype “AML AMC AMP CEF FOX” are similar to that of high cephalosporinases (Table 4).
According to the classification of Bush and Jacoby [40], there are several families of betalactamase enzymes, which differed according to the resistance phenotypes to beta lactam. In our study, beta lactamase enzymes most likely to be implicated in resistance to beta lactam antibiotics are those in Group 1 cephalosporinases, consisting of AmpC enzyme of the family of CMY enzymes (of CMY1 CMY 50) and enzyme FOX-1 and group 2 [subgroup 2b lactamases hydrolyzing penicillin and cephalosporins the latest generation] include TEM-1 enzyme, TEM-2 and SHV-1 and [under 2br groups resistant to clavulanic acid] consisting of TEM-30 and SHV-10 enzymes as well as [in group 2c and 2d hydrolyzing carbenicillin (CAR), the oxacyllines (OXA) and cloxacillines (CLX)] whose enzymes are the PSE-1, CARB-3, the RTG-4, the OXA-1 and OXA-10 [45].
Bush et al. [46] reported that AmpC enzymes are frequently located on the chromosome and are specific to Enterobacteria, contrary to the FOX-1 enzyme, localized on the plasmid specific to E. coli, Pseudomonas aeruginosa and Klbesiella pneumoniae, as to TEM1, TEM2, SHV-1, TEM-30, HVS-10, PSE-1, 3-CARB, RTG-4, OXA-1 and OXA-10 are also located on the plasmid and are specific to Enterobacteriaceae.
According to Jacoby [47], AmpC beta lactamases are inducible and can be expressed at high levels by mutation. Overexpression confers resistance to extended-spectrum cephalosporins including cefotaxime, ceftazidime, ceftriaxone, which is a particular problem due to Enterobacter aerogenes, and Enterobacter cloacae infections, where an initially isolate sensitive to these agents can become a resistant therapy.
Different resistances to aminoglycosides were observed in Table 4. According to Nikaido [38], the aminoglycosides can be inactivated by phosphoryltransférase (APH), the acetyltransferase (AAC) or nucleotidyltransferase (ANT). In this study, the enzymes were distinguished according to the classification of Becker and Cooper [48] and Morosini et al. [49] who reported the different phenotypes of resistance to aminoglycosides and the corresponding enzymes. Accordingly it was found that the enzymes which may be implicated in resistance to aminoglycosides in this study are likely to be acetyltransferases [AAC (6), AAC (3) and AAC (1)].
The results of this study identified that only Spectinomycin (SPT) is the appropriate antibiotic administered in the case of infections caused by these bacteria. The strains examined can carry several resistance genes. In fact, in accordance with Leotard and Negrin [50], the genes of resistance to different families of antibiotics are described as present on the same plasmid, representing an effective way of disseminating several related mechanisms. This spread of MDR and the spread of resistance genes are linked to the existence of mobile genetic elements between bacteria of the same species or different species, and the existence of genetic structures with many resistance genes within the same strain [51-56].
The results of this study highlighted the poor hygienic level of minced beef marketed in Constantine province making it unhealthy with evidenced risk for human consumption. These results prove the importance of raw food as a potential reservoir of bacteria that can be transferred to humans and result in a public health problem. In fact, the application of good hygiene practices all along the food chain, staff training and sensitization and the increase of the consumer’s awareness (to avoid consumption of raw and undercooked meat) may help in the reduction of food poisoning cases in our region. Furthermore, this study demonstrated the presence of MDR and possible presence of resistant genes for beta-lactamase. Any type of antimicrobial use, either for human or for animal purposes, might lead to the promotion and dissemination of resistant bacteria and genes. The accurate use of antibiotics in for animals health and in human life are essential to further control the emergence of antibiotic resistance.
- Soepranianondo K, Wardhana DK (2019) Analysis of bacterial contamination and antibiotic residue of beef meat from city slaughterhouses in East Java Province, Indonesia. Veterinary World 12: 243.
- Zerabruk K, Retta N, Muleta D, Tefera AT (2019) Assessment of microbiological safety and quality of minced meat and meat contact surfaces in selected butcher shops of Addis Ababa, Ethiopia. Journal of Food Quality.
- Li Y, Zhuang S, Mustapha A (2005) Application of a multiplex PCR for the simultaneous detection of Escherichia coli O157:H7, Salmonella and Shigella in raw and ready-to-eat meat products.Meat Sci71: 402-406. [Crossref]
- Petternel C, Galler H, Zarfel G, Luxner J, Haas D, et al. (2014) Isolation and characterization of multidrug-resistant bacteria from minced meat in Austria.Food Microbiol44: 41-46. [Crossref]
- Rašeta M, Mrdović B, Janković V, Bečkei Z, Lakićević B, et al. (2017) Prevalence and antibiotic resistance of Salmonella spp. in meat products, meat preparations and minced meat. Earth and Environmental Science 85: 012028.
- Boudechicha HR, Nasri I, Bennaceur Z, Sellama M, Hafid K, et al. (2017) Microbiological changes during the preparation steps of Khliaa Ezir: a traditional cured meat product of Algeria. Integr Food Nutr Metab 4: 1-5.
- CA-SFM. EUCAST (2019) Comité de l’antibiogramme de la Société Française de Microbiologie, Recommandations 2019.V.1.O Janvier, pp: 144.
- Farrag HA, El-Zawahry YA, El-Aziz SMA, Nada HM (2008) Detection of extended-spectrum B-lactamase producers among gram-negative bacilli isolated from clinical samples. Egyptian Journal of Medical Microbiology 17: 389-404.
- Iqbal R, Ikram N, Shoaib M (2017) Phenotypic cofirmatory disc diffusion test (PCDDT), double disc synergy test (DDST), E-test OS diagnostic tool for detection of extended spectrum beta lactamase (ESΒL) producing Uropathogens. J Appl Biotechnol Bioeng 3: 344-349.
- Bouzid R, Guemour D, Zidane K, Aggad H, Bendella A, et al. (2015) Hygienic quality of minced meat retailed in western Algeria. Journal of Virology and Microbiology 2015: C1-9.
- Oumokhtar B, Berrada H, Ameur N, El Fakir S (2008) Analyse microbiologique de la viande hachée bovine commercialisé à Fès, Maroc. Les Technol de Laboratoire 12: 4-10.
- Tassew H, Abdissa A, Beyene G, Gebre-Selassie S (2010) Microbial flora and food borne pathogens on minced meat and their susceptibility to antimicrobial agents. Ethiopian Journal of Health Sciences 20: 137-143.
- El-Gendy NM, Ibrahim HA, Al-Shabasy NA, Samaha IA (2014) Enterobacteriaceae in beef products from retail outlets in Alexandria. Alexandria Journal of Veterinary Sciences 41: 80-86.
- Yusuf AB, Gulumbe BH, Aliyu B, Kalgo ZM (2019) Bacteriological assessment of fresh beef sold in Birnin Kebbi Central Market, Kebbi State, Nigeria. International Journal of Medical Research and Health Sciences 8: 127-131.
- Joffin C, Joffin JN (1999) Microbiologie Alimentaire. Edition CRDP. Bordeaux, 5ème éd. Collection Biologie Technique, 211-212.
- JORA (2017) Journal Officiel de la République Algérienne Démocratique et Populaire N° 39. Critères microbiologiques relatifs à certaines denrées alimentaires. Décret exécutif n° 15-172.
- Lasta JA, Rodríguez R, Zanelli M, Margaría CA (1992) Bacterial count from bovine carcasses as an indicator of hygiene at slaughtering places: A proposal for sampling. Journal of Food Protection 55: 271-278.
- Zweifel C, Fischer R, Stephan R (2008) Microbiological contamination of pig and cattle carcasses in different small-scale Swiss abattoirs. Meat Science 78: 225-231.
- Cohen N, Ennaji H, Hassa M, Karib H (2006) The bacterial quality of red meat and offal in Casablanca (Morocco). Molecular Nutrition & Food Research 50: 557-562.
- Blood RM, Curtis GDW (1995) Media for ‘total’ Enterobacteriaceae, coliforms and Escherichia coli. International Journal of Food Microbiology 26: 93-115.
- Sousa CPD (2008) The impact of food manufacturing practices on food borne diseases. Brazilian Archives of Biology and Technology 51: 615-623.
- Wanyenya I, Muyanja C, Nasinyama GW (2004) Kitchen practices used in handling broiler chickens and survival of Campylobacter spp. on cutting surfaces in Kampala, Uganda. Journal of Food Protection 67: 1957-1960.
- Hart CA, Winstanley C (2001) What makes a pathogen? Microbiology Today 28: 4-6.
- Regab WS, Hassan EAB, Al-Geddawy MA, Albie AA (2016) Bacteriological quality of some meat products in the Egyptian retail markets. Assiut J Agric Sci 47: 422-429.
- Hassan NA, Farooqui A, Khan A, Khan AY, Kazmi SU (2010) Microbial contamination of raw meat and its environment in retail shops in Karachi, Pakistan. Journal of Infection in Developing Countries 4: 382-388.
- Rasool SA, Ahmad AF, Khan S, Wahab A (2003) Plasmid borne antibiotic resistance factors among indigenous Klebsiella. Pak J Bot 35: 243-248.
- Diab AM, Abdel Aziz MH, Selim SA (2002) Plasmid-encoded transferable antibiotic resistance in gram-negative bacteria, isolated from drinking water in Ismaillia city. Pakistan Journal of Biological Sciences 5: 774-779.
- Thavasi R, Aparnadevi K, Jayalakshmi S, Balasubramanian T (2007) Plasmid mediated antibiotic resistance in marine bacteria. Journal of Environmental Biology 28: 617.
- Carattoli A (2009) Resistance plasmid families in Enterobacteriaceae. Antimicrobial Agents and Chemotherapy 53: 2227-2238.
- DebMandal M, Mandal S, Pal NK (2011) Antibiotic resistance prevalence and pattern in environmental bacterial isolates. The Open Antimicrobial Agents Journal 3: 45-52.
- Nsofor CA, Iroegbu CU (2013) Plasmid profile of antibiotic resistant Escherichia coli isolated from domestic animals in South-East Nigeria. Journal of Cell and Animal Biology 7: 109-115.
- Reinthaler FF, Feierl G, Galler H, Haas D, Leitner E, et al. (2010) ESBL-producing E. coli in Austrian sewage sludge.Water Res44: 1981-1985. [Crossref]
- Gandolfi I, Franzetti A, Bertolini V, Gaspari E, Bestetti G (2011) Antibiotic resistance in bacteria associated with coarse atmospheric particulate matter in an urban area. Journal of Applied Microbiology 110: 1612-1620.
- Doyle ME (2015) Multidrug-resistant pathogens in the food supply. Foodborne Pathogens and Disease 12: 261-279.
- Newell DG, Koopmans M, Verhoef L, Duizer E, Aidara-Kane A, et al. (2010) Food-borne diseases — The challenges of 20 years ago still persist while new ones continue to emerge. International Journal of Food Microbiology 139: S3-S15.
- Willems E, Cartuyvels R, Magerman, K, Raymaekers M, Verhaegen J (2013) Comparison of different phenotypic assays for the detection of extended-spectrum β-lactamase production by inducible AmpC-producing Gram-negative bacilli. European Journal of Clinical Microbiology & Infectious Diseases 32: 549-555.
- Pares S, Mouz N, Petillot Y, Hakenbeck R, Dideberg O (1996) X-ray structure of Streptococcus pneumoniae PBP2x, a primary penicillin target enzyme. Nature Structural Biology 3: 284.
- Nikaido H (2009) Multidrug resistance in bacteria.Annu Rev Biochem78: 119-146. [Crossref]
- Falagas ME, Karageorgopoulos DE (2009) Extended-spectrum β-lactamase-producing organisms. Journal of Hospital Infection 73: 345-354.
- Bush K, Jacoby GA (2010) Updated functional classification of β-lactamases. Antimicrobial Agents and Chemotherapy 54: 969-976.
- Ruppé E, Bidet P, Verdet C, Arlet G, Bingen E (2006) First detection of the Ambler class C 1 AmpC β-lactamase in Citrobacter freundii by a new, simple double-disk synergy test. Journal of Clinical Microbiology 44: 4204-4207.
- Carter MW, Oakton KJ, Warner M, Livermore DM (2000) Detection of extended-spectrum β-lactamases in Klebsiellae with the Oxoid combination disk method. Journal of Clinical Microbiology 38: 4228-4232.
- Chen CY, Wu KM, Chang YC, Chang CH, Tsai HC, et al. (2003) Comparative genome analysis of Vibrio vulnificus, a marine pathogen.Genome Res13: 2577-2587. [Crossref]
- Courvalin P, Philippon A (1989) Mécanismes biochimiques de la résistance bactérienne aux agents antibactériens. Bactériologie médicale. Paris: Flammarion, 332-55.
- Dib AL (2014) Evaluation de la contamination microbienne des produits de la mer. Thèse pour l’obtention de doctorat en Science. Option, Hygiène et sécurité alimentaire. Université Consantine 1. Institut des Sciences Vétérinaires. 206p.
- Bush K, Jacoby GA, Medeiros AA (1995) A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrobial Agents and Chemotherapy 39: 1211.
- Jacoby GA (2009) AmpC β-lactamases. Clinical Microbiology Reviews 22: 161-182.
- Becker B, Cooper MA (2012) Aminoglycoside antibiotics in the 21st century. ACS Chemical Biology 8: 105-115.
- Morosini MI, Cercenado E, Ardanuy C, Torres C (2012) Detección fenotípica de mecanismos de resistencia en microorganismos grampositivos. Enfermedades Infecciosas y Microbiología Clínica 30: 325-332.
- Leotard S, Negrin N (2010) Épidémiologie des entérobactéries sécrétrices de bêta-lactamases à spectre étendu (E-BLSE) au centre hospitalier de Grasse (2005-2008). Pathologie Biologie 58: 35-38.
- Skurnik D, Andremont A (2006) Antibiothérapie sélectionnante. De la théorie à la pratique. Réanimation 15: 198-204.
- EFSA Panel on Biological Hazards (BIOHAZ) (2014) Scientific opinion on the public health risks related to the maintenance of the cold chain during storage and transport of meat. EFSA Journal 12: 3783.
- Fosse J, Cappelier JM, Laroche M, Fradin N, Giraudet K, Magras C (2006) Viandes bovines: une analyse des dangers biologiques pour le consommateur appliquée à l’abattoir. Rencontre Recherche Ruminants 13: 411-414.
- Kımıran E, Saglam D, Ozer D, Ozcelık E (2014) Microbiological quality of minced meat samples marketed in Istanbul. Yüzüncü yıl Üniversitesi Veteriner Fakültesi Dergisi 25: 67-70.
- Salifou CFA, Youssao AKI, Ahounou GS, Tougan PU, Farougou S, et al. (2013) Critères d’appréciation et facteurs de variation des caractéristiques de la carcasse et de qualité de la viande bovine. Annales de Médecine Vétérinaire 157: 27-42.
- Tegegne M, Ashenafi M (1998) Microbial load and incidence of Salmonella spp. inKitfo', a traditional Ethiopian spiced, minced meat dish. Ethiopian Journal of Health Development 12: 135-140.

