Abstract
[GADV]-protein world hypothesis or GADV hypothesis on the origin of life was obtained from the study on the way how an entirely new (EntNew) gene has been created in extant microorganisms. Therefore, the GADV hypothesis is intimately related to creation of EntNew gene/protein on the present Earth. In this short note, I will introduce possible strategies for application of EntNew protein created in GC-rich microorganism to medical or clinical research.
Introduction
I have proposed [GADV]-protein world hypothesis or GADV hypothesis on the origin of life, advocating that life emerged from [GADV]-protein world, which was formed by pseudo-replication of [GADV]-protein or aggregates of [GADV]-peptides [1-3], but not from RNA world [4]. [GADV] means four amino acids, Gly [G], Ala [A], Asp [D] and Val [V].
Research on the origin of life undoubtedly belongs to one of basic researches. Therefore, many people may consider that results obtained by the research could not be applied to an applied science as medical or clinical research. However, it is also well known that causes of many genetic diseases have been well understood with a knowledge obtained through basic researches, as molecular biology and protein chemistry (see, for example, Ref. No. [5]). Then, I will introduce an idea in this short note, about the way how one of the results, or GC-NSF(a) hypothesis on the origin of gene [6,7], which was obtained through the life-origin research could be applied to clinical research, actually, to synthesis of a medicine, which could be useful for medical treatments, and to treatments of a protein causing disease.
How was GADV hypothesis obtained?
I started the life-origin research from a study for understanding the way how an entirely new (EntNew) gene, which did not previously exist is formed [1-3]. Therefore, I did not intend to solve the riddle on the origin of life from the start. And, I fortunately reached one idea, GC-NSF(a) hypothesis on the origin of EntNew gene, suggesting that EntNew gene has been produced from non-stop frame on antisense strand (NSF(a)) of, not AT-rich, but GC-rich microbial gene [6,7] (Figure 1).
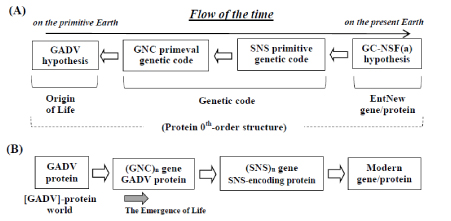
Figure 1(A). The progress, which I reached GADV hypothesis. The study was initiated from search of a field of entirely new (EntNew) gene/protein, resulted in proposition of GC-NSF(a) hypothesis, suggesting that EntNew gene is formed from nonstop frame on antisense sequence of GC-rich gene. Successively, I reachedGNC-SNS primitive genetic code hypothesis, which triggered discovery of the GADV hypothesis. All the three hypotheses were obtained based on protein 0th-order structure. Bold white arrows indicate the direction of the study. Figure1(B) The emergence of life and the evolutionary process to modern life system, deduced from GADV hypothesis. The emergence of life was founded by formation of [GADV]-protein world, in which nucleotides and oligonucleotides were produced. Life emerged after a necessary number of (GNC)n genes and [GADV]-proteins were formed. Bold white arrows indicate the direction of evolutionary process from [GADV]-protein world to the modern gene/protein. Dark bold arrow indicates a deduced time point, at when the first life emerged.
Successively, I confirmed that base compositions at three codon positions of both highly GC-rich genes (65~70%) and their GC-NSF(a) sequences are roughly SNS or [(G/C)N(C/G)] [1-3,6], and that the computer-generated SNS code encoding 10 amino acids satisfies the six conditions (hydrophobicity/hydrophilicity, a(α)-helix, b(β)-sheet and turn/coil formabilities, acidic amino acid composition and basic amino acid composition) when the compositions of G and C at the first codon position are at around 55% and 45%, respectively, and when four bases were contained at about one-fourth each at the second codon position. Based on the facts, I proposed SNS primitive genetic code hypothesis [8] (Figure 1).
Furthermore, I thought of that the SNS code is still too complex to be the most primitive genetic code, since the code is composed of 16 codons and 10 amino acids. Then, I searched for simpler code than SNS code and found out that [GADV]-protein encoded by GNC code satisfies the four conditions (hydrophobicity/hydrophilicity, a(α)-helix, b(β)-sheet and turn/coil formabilities), when about equal amounts of [GADV]-amino acids are contained in a protein. Thus, we got GNC-SNS primitive genetic code hypothesis (Figure 1 (A)) [9].
If the GNC code was the first genetic code, the simplest set of amino acids, which were used to produce functional water-soluble globular protein on the primitive Earth, should be [GADV]-amino acids. Taking these into consideration, I proposed the GADV hypothesis on the origin of life about 15 years ago (Figure 1(A)), suggesting that life emerged from [GADV]-protein world, which was formed by pseudo-replication of [GADV]-protein (Figure 1 (B)) [1-3].
How was EntNew gene/protein produced?
It is well known that homologous gene/protein has been formed from genetic sequence on sense strand of a previously existing gene after duplication. The idea was proposed by S. Ohno as gene duplication theory [10]. On the contrary, it was totally unknown about 30 years ago, how EntNew gene/protein was created. However, I now consider that EntNew gene/proteins were generated from (GNC)n, (SNS)n and GC-NSF(a) sequences on antisense strand of (GNC)n, (SNS)nand GC-rich genes in GNC, SNS and universal genetic code era, respectively (Figure 1(B)) [3]. Take notice of that (GNC)nand (SNS)nsequence are also highly GC-rich as about 83%.
Possible application of the life-origin research or GADV hypothesis to medical treatments.
Next, I would like to provide a new strategy about the way how a knowledge obtained during the study on GADV hypothesis can be applied to clinical research [Figure 2].
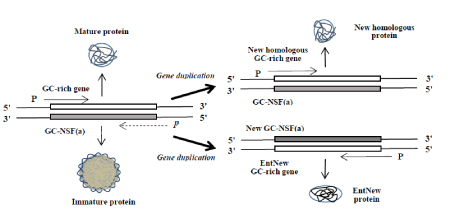
Figure 2.Formation of new homologous and entirely new (EntNew) genes/proteins. New homologous gene/protein is produced from sense sequence of a parental gene, after gene duplication as proposed by S. Ohno [10]. On the other hand, EntNew gene/protein is created from nonstop frame on antisence strand of GC-rich gene (GC-NSF(a)) also after gene duplication, as suggested by GC-NSF(a) hypothesis, which I proposed [6,7]. Wavy lines on the surface of and thin lines in immature protein indicate flexible structure of the protein. Capital letters, P, and a small, italic letter, p, indicate a promoter with sufficiently high transcription initiation activity and a cryptic promoter with a low function for the initiation, respectively.
Protein functions used in extant organisms extend to extraordinary wide range, as easily understood from that organisms living in extreme circumstances, such as at an extremely high or low temperature, and in highly acidic or basic condition, have been discovered. Creation of EntNew gene/protein necessary to live in such extreme conditions could have made it possible for those organisms to adapt to the respective circumstances. In addition, amino acid sequence diversity of a small protein composed of only 100 amino acid residues reaches 20100 = ~ 10130. These mean that there are many possibilities to create EntNew protein with a new function, which is not used in any organisms inhabiting on the present Earth.
Then, I will provide a new strategy with EntNew gene/protein as follows. First one is to detoxify a toxic compound by dimerization or degradation (Figure 3(A)) of the toxin with EntNew enzyme, which is produced according to GC-NSF(a) hypothesis. Such kind of EntNew enzyme could be created in a bacterium carrying GC-rich genome, such as Pseudomonas aeruginosa (GC content of the genome = ~67%) through repeated cultivation of the bacterium in the medium containing a limited amount of the toxin. Second one is to detoxify a protein causing disease by complex formation between the toxic protein and EntNew protein or by degradation of the protein with EntNew protein, as shown in Figure 3(B).
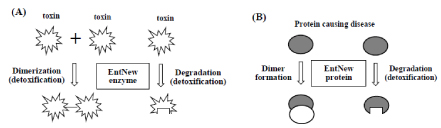
Figure 3.Possible strategy for detoxifying a toxic chemical compound (A) and a protein causing a disease (B) with EntNew enzyme or protein. Such an enzyme or protein could be created in a bacterium carrying GC-rich genome through repeated cultivation of the bacterium in a medium containing at a limited amount of the toxin or the protein. The reason is because the bacterium could grow faster than before, if EntNew enzyme or protein for detoxifying the toxin or the protein was produced in the cells.
Only simple examples for application of EntNew protein (enzyme) to medical treatments are given in this short note. However, EntNew protein created according to GC-NSF(a) hypothesis could be applied to many fields, such as synthesis of new medicine, decontamination of organic pollutes and chemical industries etc., because amino acid sequence diversity is extraordinary wide as ~10130, as described above, so that useful proteins with a novel catalytic activity could be created, if necessary.
Acknowledgments
I am grateful to Tadashi Oishi (G & L Kyosei Institute, Emeritus professor of Nara Women’s University) for the encouragement throughout my research on the GADV hypothesis on the origin of life.
Conflicts of Interest
The author declares no conflict of interest.
References
- Ikehara K1 (2002) Origins of gene, genetic code, protein and life: comprehensive view of life systems from a GNC-SNS primitive genetic code hypothesis. J Biosci27: 165-186. [Crossref]
- Ikehara K (2005) Possible steps to the emergence of life: The [GADV]-protein world hypothesis. Chem Rec5: 107–118.
- Ikehara K (2016)“GADV hypothesis on the origin of life –Life emerged in this way-” LAPLAMBERT Academic Publishing, Saarbrucken, Germany.
- GilbertW (1986)The RNA world. Nature319: 618.
- IkeharaK(2011) Advances in the Study of Genetic Disorders.InTech-Open Access Publisher, Rijeka, Croatia.
- Ikehara K, Amada F, Yoshida S,Mikata Y, Tanaka A (1996) A possible origin of newly-born bacterial genes: Signi?cance of GC-rich nonstop frame on antisense strand. Nucleic Acids Res24: 4249–4255.
- Ikehara K (2003) Simulation of gene evolution: Evidence for GC-NSF(a) hypothesis on the origin of genes. Viva Origino31: 201–215.
- Ikehara K, Yoshida 2021 Copyright OAT. All rights reserv of the genetic code.Viva Origino26: 301-310.
- Ikehara K, Omori Y, Arai R, Hirose A (2002) A novel theory on the origin of the genetic code: a GNC-SNS hypothesis. J MolEvol54: 530-538.[Crossref]
- Ohno S (1970) “Evolution by gene duplication”. Springer, Heiderberg, Germany.



