Background: Sepsis requires an appropriate and quick treatment based on a fast determination of pathogen and antimicrobial susceptibility. It is much needed to explore a new protocol to shorten the turnaround time of currently standard process (SP) for pathogen identification (PI) and antimicrobial susceptibility tests (AST) used in most of hospitals without addition expensive devices.
Materials and methods: This study reported a new expedited process (NEP) that directly took 10 ml positive culture media from BD BACTEC bottles, lysed blood cells, obtained the bacterial pellet and directly processed for PI and AST. A side-by-side comparison between NEP and SP was carried out in 20 patients.
Results: While NEP shorten the turnaround time for at least 1 day, the side-by-side comparison showed: (a) NEP yielded the PI results identical to SP; (b) in 305 paired AST:297(97.38%)were identical, only 4 (1.31%) had major error (Resistant to Sensitive or Sensitive to Resistant) and 4 (1.31%) had minor error (Intermediate to Sensitive or Resistant). Advantages of NEP are: (1) compared with currently used SP,NEP does not need overnight dish culture for colonies, saving manpower and materials, and yielding the reports of PI and AST at least 1day earlier than SP; (2) compared to other newly developed rapid-techniques, NEP utilizes SP devices without addition of new expensive devices and reagents, which is acceptable easily by all current SP users.
Conclusion: NEP is acceptable to replace SP with an advantage of shortening at least 1day turnaround time for PI and AST,which will have a significant impact on the treatment decision-making and outcome of patients with systemic infection.
pathogen identification, antimicrobial susceptibility, early diagnosis of sepsis
Abbreviations
SP: Standard Process; NEP: New Expedited Process; McF: Mcfarland turbidimetric unit (bacterial concentration); PI: Pathogen Identification; AST: Antimicrobial Susceptibility Tests
Sepsis,up to ~30% death [ Obviously, the early turnaround times for PI and AST are critical for rescuing life [
At present, in most of hospitals use the , which takes at least 2-3 days to report the pathogen stain and antimicrobial susceptibility []: on day one, to culture blood in BD ;on day two, if culture is positive , the pathogen is inoculated to the dish for colonies to grow overnight; on day three, all grown colonies are collected,justified to concentrationof 0.5 McF for PI and using and AST[
Great efforts have been made to develop various instrument-assisted PI and AST systems to shorten the turnaround time to in hours rather than SP in days [16-17]. Recently, the multiplex polymerase chain reaction–based rapid diagnostic tests, such as Film Array Blood Culture Identification (FA-BCID), is developed to detect pathogens and antimicrobial resistance genes for rapid reports in about one hour, however, an initial blood culture is still required for a positive microbial growth [18]. A matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) is also used for a rapid report [19]. All these new techniques require expensive devices, which are not available in most of hospitals [12]. Therefore, it badly need to utilize SP currently used devices and systems by changing protocol to shorten the turnaround time of PI and AST before most of hospitals could be equipped with expensive FA-BCID or MALDI-TOF MS.
After carefully observation of the currently used SP, we found that the inoculation of bacteria from bottle to dish for further cultured overnight could be omitted, due to: (1) instead of picking up single colony, all the colonies grown overnight in the whole dish were harvested together, pooled into the same tube and adjusted to 0.5 McF for PI and AST; (2) in almost all the cases, the pathogen report from VITEK 2 reader of whole dish is single bacterial strain; and (3) very few culture dish grows more than one strain of bacteria. Therefore, we believe that directly processing the bacterial positive media from the BD BACTEC bottle is likely to yield the same results of PI and AST as these from the re-culture in dish overnight. To prove if NEP omits the step of overnight dish culture could yield the same results as SP (Figure 1), we performed this study with a goal of shorten the turnaround time for at least one day without changing any used device and associated reagents, which could be welcome by physicians and patients for its aiding in the early and accurate treatment of sepsis.
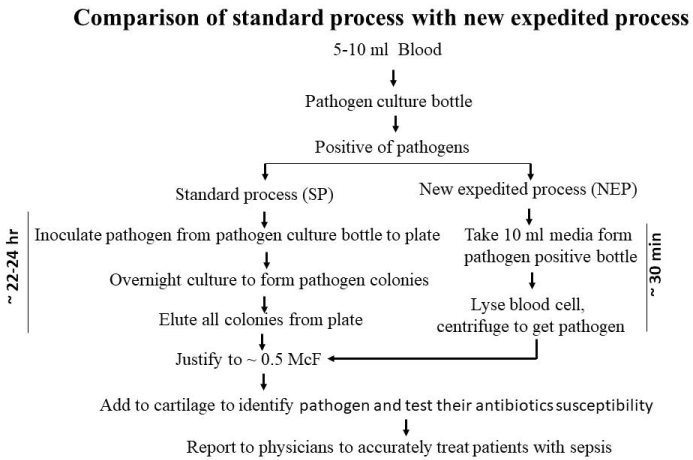
Figure 1. Comparison of processes between SP and NEP: Broth taken from a blood culture positive bottle, either inoculated on to dish for overnight culture of colonies for SP or transferred 10 ml broth to a 50 ml tube for lysis of blood cellular portion for NEP, followed by making 0.5 McF for PI and AST with VITEK 2 device and its associated cards. NEP omitted the dish re-culture step, which yielded results about 22-24 h earlier than SP.
Sample collection
Patients with 2 of 4 following clinical symptoms were highly suspected as sepsis [16]: (1) a body temperature of > 38° C or < 36° C; (2) a heart rate > 90 beats per min; (3) a respiratory rate > 20 breaths per minute or an arterial CO2 pressure of < 32 mm Hg; and (4) white blood cell counts of > 12,000 cells/ L or < 4000 cells/ L or > 10% immature forms. Then, their blood samples were collected and cultured in BD BACTEC bottle (Becton Dickinson and Company Sparks, MD, USA 21157) were performed for 7 day [17-18]. Twenty cultured blood samples out of about 200 samples from hospitalized patients in 4 months of year 2015 were positive for either gram-negative or gram-positive bacteria in 12-72 hours after culture. The positive samples in BD BACTEC bottle were further processed according to procedures of either routine SP or our NEP. The sample collection was approved by the institutional review board of First Affiliated Hospital of Fujian Medical University (protocol# 2015-084).
Standard process (SP)
As showed in figure 1, the positive samples from BD BACTEC bottle were inoculated in to bacterial culture dish for overnight culture. Next day (about 20-22 hours later), all colonies grown in the dish were eluted with saline and justified to the final bacterial concentration of 0.5 McF with Mcfarland turbidimetric device (DensiCHEK plus, Biomerieux, Andover, Massachusetts, USA) , then used VITEK 2 system and its cards for PI {Gram-Negative (or Positive) Identification cards} and AST {Gram-Negative (or Positive) AST cards}.
New expedited process (NEP)
As showed in figure 1, 10 ml bacterial positive culture media from BD BACTEC bottle was taken out with a 20 ml of sterilized syringe, placed in 50 ml sterilized tube, added 35 ml of sterile water and then 5 ml of 10 × filtered blood cell lysis buffer (NH4Cl 83 g; NaHCO3 10 g; EDTA 370 mg; addition of sterile water to 500 ml), sharking the tube gently by a rotator (60 rpn) for about 8-10 min until the solution in the tube was completely transparent, then centrifuged (1500 g, 10 min) to obtain the bacterial pellet. After wash with saline once, the bacterial pellet was re-suspend in saline and justified to 0.5 McF followed by using VITEK 2 devices and cards for PI and AST as done in SP above.
Pathogen identification
The same volume (0.5 ml) of 0.5 McF each from SP (colonies harvested from overnight culture dish) and NEP (10 ml broth directly taken from positive BD BACTEC bottle, lysis of blood cell for pellet of bacteria) from each of 20 patients was transferred separately into Gram-Negative (or Positive) Identification cards specially made by bioMerieux Inc and inserted into VITEK 2 device (bioMerieux Inc, 595 Anglum Road, Hazelwood, MO 63042,USA) for PI. The results of bacterial strains were compared side-by-side between SP and NEP.
Antimicrobial susceptibility tests (AST)
According to the result of Gram stain, the same volume (0.5 ml) of 0.5 McF each from SP and NEP bacterial preparation of same patient was also transferred separately into either Gram-positive test kit (VITEK 2AST-GP67, including 21 drug tests) or Gram-negative test kit (VITEK 2AST-GN09, including 22 drug tests) and then inserted into VITEK 2 device for the AST. The results of AST were compared side-by-side between SP and NEP.
Statistical analysis
The McNemar-bowker paired chi-square test was used to test the differences between the two procedures used in the same sample, X2 = 2.000, p < 0.05 was statistically significant difference. The kappa consistency test was used to examine the consistency of the two procedures. When Kappa was > 0.9, indicating that the consistency was excellent.
The pathogen identification is identical between SP and NEP
As shown in figure 2A, in 20 blood culture positive cases, media from each positive culture bottle was taken either inoculating into culture dish for isolates as SP or 10 ml subjected to NEP. The PI results from two procedures was identical, i.e. 100% category agreement in all pathogen systemically invaded in 20 patients, including Staphylococcus aureus (n = 7), Klebsiella pneumoniae ssp pneumoni (n = 6), Escherichia coli (n = 3), Acinetobacter baumannii (n = 1), Enterococcus faecium (n = 1), Pseudomonas aeruginosa (n = 1), Enterobacter aerogenes (n = 1). Data demonstrated that: (1) NEP yielded the PI not only identical to SP, but also about 24 hours earlier than SP; and (2) the invaded genus was monomicrobial in all these 20 cases.
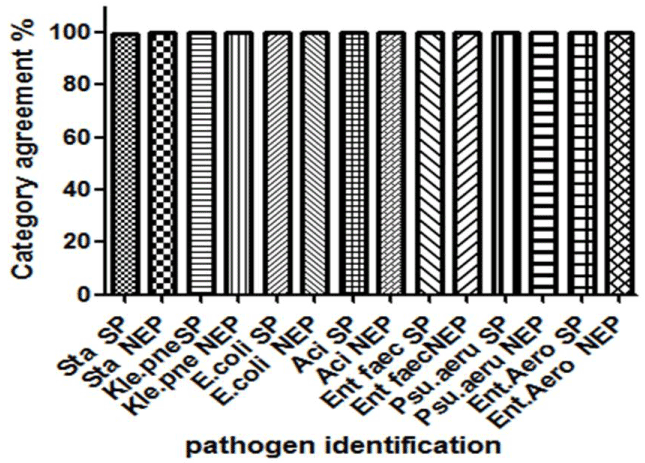
Figure 2A. The pathogen identification is identical between SP and NEP: In 20 patients tested for genus, SP and NEP yielded identical pathogens (Mcnemar-bowker paired chi-square test, P > 0.5): 7 cases with Staphylococcus aureus, 6 Klebsiella pneumoniae, 3 Escherichia coli, 1 Acinetobacter baumannii, 1 Enterococcus faecium, 1 Pseudomonas aeruginosa and 1 Enterobacter aerogenes. All 20 patients were systemically infected with mono-microbial.
The results of AST are largely agreed between SP and NEP
The AST includes the following antibiotics: gentamicin (GM), levofloxacin (LEV), aztreonam (ATM), piperacillin (PIP), nitrofurantoin (FT),trimethoprim/sulfa (SXT), erythroprim/sulfa (E), ciprofloxacin (CIP), amikacin (AN), imipenem (IPM), clindamycin (CM), quinupristin/dalfopristin (QDA), moxifloxacin (MXF), tetracycline (TE), vancomycin (VA), ampicillin (AM), ampicillin-sulbactam (SAM),tobramycin (TM), cefotetan (CTT), ceftazidime (CAZ), ceftriaxone (CRO), aztreonam cefepime (FEP), piperacillin (TZP), piperacillin (PIP), tigecycline (TGC), oxacillin (OX1),cefuroxime (CXM), linezolid (LNZ), penicillin-g/ benzylpenicillin (P), rifampin (RA), and meropenem (MEM).
Compared SP with NEP, as shown in figure 2B, among total 305 paired AST (SP v.s. NEP), 297 paired results (297/305, 97.38%) were identical. There were 4 tests (4/305, 1.31%) with minor errors, sensitivity from intermediate to sensitive or resistant (I to S or I to R), shown 1 with Staphylococcus aureus (Table 1), 1 with Escherichia coli (Table 3), 2 with Enterobacter aerogenes (Table 7). There were 4 tests (4/305, 1.31%) with major errors, such as sensitivity from resistant or sensitive (R or S) to sensitive or resistant (S or R), shown 3 with Staphylococcus aureus (Table 1), and 1 with Pseudomonas aeruginosa (Table 6).
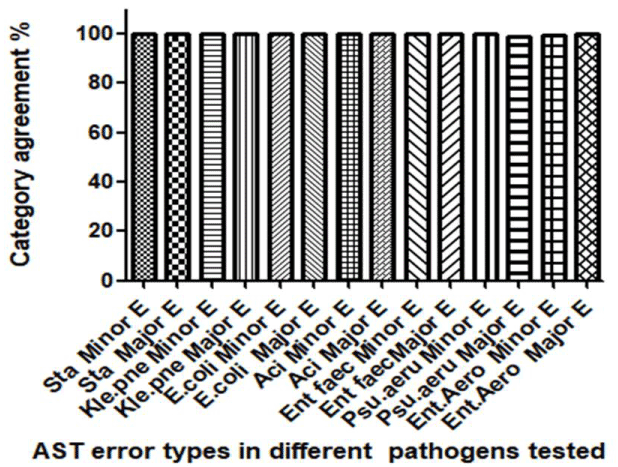
Figure 2B. The antimicrobial susceptibility is largely agreed between SP and NEP: Totally, 305AST were performed in 20 cases with 7 pathogens. 297 out of 305 (97.38%) AST results were identical, 4 with minor error (1.31%) and 4 with major error (1.31%, the kappa consistency test, P > 0.9).
Table 1-7 demonstrated the detail of each paired AST with the names of antibiotics. The AST agreement rate in total 305 tests was 97.38% with 4 (1.31%) minor errors (I to S or R) and 4 (1.31%) major errors (S to R or R to S). Specifically, as shown in figure 2C, among 31 antibiotics tested in 305 paired samples, CA, RA, GM and CAZ each had one major error, PIP had two minor errors, LNZ and IPM each had one minor error. The data suggest that NEP yields very low error rate, therefore, it is acceptable as compared to SP with advantage of one day earlier reports of AST to physicians.
Table 1. Antibiotics susceptibility tests for Staphylococcus with two procedures*
|
Case 1 |
Case 2 |
Case 3 |
Case 4 |
Case 5 |
Case 6 |
Case 7 |
SP |
NEP |
SP |
NEP |
SP |
NEP |
SP |
NEP |
SP |
NEP |
SP |
NEP |
SP |
NEP |
Pathogen identification |
Sta. homi
|
Sta.homi |
Sta.aure |
Sta.aure |
Sta.aure |
Sta.aure |
Sta.aure |
Sta.aure |
Sta..aure |
Sta..aure |
Sta..aure |
Sta..aure |
Staphylo |
Staphylo |
Oxacillin |
S |
S |
R |
R |
R |
R |
S |
S |
S |
S |
|
|
R |
R |
Nitrofurantoin |
S |
S |
S |
S |
|
|
|
|
S |
S |
|
|
R |
R |
Trimethoprim/ sulfa |
S |
S |
R |
R |
|
|
|
|
S |
S |
|
|
R |
R |
Erythroprim/sulfa |
|
|
|
|
R |
R |
S |
S |
S |
S |
|
|
R |
R |
Ciprofloxacin |
R |
R |
R |
R |
|
|
S |
S |
S |
S |
S |
S |
S |
S |
Clindamycin |
S |
S |
R |
R |
S |
R |
S |
S |
S |
S |
S |
S |
S |
S |
Quinupristin/dalfopristin |
R |
R |
R |
R |
|
|
S |
S |
S |
S |
S |
S |
|
|
Rifampin |
S |
S |
R |
S |
|
|
S |
S |
S |
S |
S |
S |
R |
R |
Moxifloxacin |
S |
S |
|
|
|
|
S |
S |
S |
S |
S |
S |
S |
S |
Penicillin-G |
|
|
|
|
|
|
|
|
|
|
|
|
S |
S |
Gentamicin |
S |
S |
S |
R |
|
|
S |
S |
S |
S |
S |
S |
S |
S |
Tetracycline |
|
|
|
|
|
|
S |
S |
S |
S |
S |
S |
I |
I |
Tigecycline |
S |
S |
S |
S |
|
|
S |
S |
S |
S |
S |
S |
R |
R |
Vancomycin |
R |
R |
S |
S |
|
|
S |
S |
|
|
S |
S |
S |
S |
Levofloxacin |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
Benzylpenicillin |
S |
S |
S |
S |
|
|
R |
R |
|
|
S |
S |
S |
S |
Linezolid |
S |
S |
I |
S |
|
|
S |
S |
|
|
S |
S |
|
|
Nitrofurantoin |
|
|
|
|
|
|
S |
S |
|
|
S |
S |
|
|
Trimethoprim/ sulfa |
S |
S |
|
|
|
|
|
|
|
|
|
|
|
|
S: Sensitive; R: Resistant; I: Intermediate; *The kappa consistency Test was used, Kappa value = 0.875, 95% CI: 0.743-0.972, Kappa > 0.9, indicating that the consistency was excellent.
Table 2. Antibiotics susceptibility tests for six cases of Klebsiella pneumoniae ssp pneumoni with two procedures*
|
Case 8 |
Case 9 |
Case 10 |
Case 11 |
Case 12 |
Case 13 |
|
SP |
NEP |
SP |
NEP |
SP |
NEP |
SP |
NEP |
SP |
NEP |
SP |
NEP |
Pathogen identification |
Kleb. pneumoni |
Kleb. pneumoni |
Kleb. pneumoni |
Kleb. pneumoni |
Kleb. pneumoni |
Kleb. pneumoni |
Kleb. pneumoni |
Kleb. pneumoni |
Kleb. pneumoni |
Kleb. pneumoni |
Kleb. pneumoni |
Kleb. pneumoni
|
Meropenem |
S |
S |
S |
S |
S |
S |
R |
R |
S |
S |
S |
S |
Piperacillin |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
Piperacillin/ Tazobactam |
S |
S |
S |
S |
S |
S |
R |
R |
S |
S |
S |
S |
Amikacin |
S |
S |
S |
S |
R |
R |
R |
R |
S |
S |
S |
S |
Ampicillin |
R |
R |
R |
R |
S |
S |
R |
R |
R |
R |
R |
R |
Ampicillin/ Sulbactam |
S |
S |
S |
S |
S |
S |
R |
R |
S |
S |
R |
R |
Aztreonam |
S |
S |
S |
S |
S |
S |
R |
R |
S |
S |
|
|
Cefepime |
S |
S |
S |
S |
S |
S |
R |
R |
|
|
S |
S |
Cefuroxime |
S |
S |
S |
S |
S |
S |
R |
|
S |
S |
S |
S |
Cefuroxime Axetil |
S |
S |
|
|
S |
S |
R |
R |
S |
S |
R |
R |
Ceftriaxone |
S |
S |
S |
S |
S |
S |
R |
R |
S |
S |
R |
R |
Ceftazidime |
S |
S |
S |
S |
S |
S |
R |
R |
S |
S |
R |
R |
Cefotetan |
S |
S |
S |
S |
S |
S |
R |
R |
S |
S |
S |
S |
Tobramycin |
S |
S |
S |
S |
S |
S |
R |
R |
S |
S |
S |
S |
Imipenem |
S |
S |
S |
S |
S |
S |
R |
R |
S |
S |
S |
S |
Levofloxacin |
S |
S |
S |
S |
I |
I |
|
|
S |
S |
S |
S |
Nitrofurantoin |
S |
S |
I |
I |
S |
S |
|
|
S |
S |
S |
S |
Trimethoprim/ sulfa |
S |
S |
S |
S |
S |
S |
R |
R |
I |
I |
I |
R |
Ciprofloxacin |
S |
S |
|
|
|
|
R |
R |
|
|
R |
R |
Gentamicin |
S |
S |
|
|
|
|
R |
R |
S |
S |
S |
S |
Cefuroxime |
|
|
S |
S |
S |
S |
|
|
|
|
|
|
S: Sensitive; R: Resistant; I: Intermediate; Kleb. pneumoni: Klebsiella pneumoniae ssp pneumoni; *The kappa consistency Test was used, Kappa = 1.000, indicating that the consistency was excellent.
Table 3. Antibiotics susceptibility tests for Escherichia coli with two procedures*
|
Case 14 |
|
Case 15 |
|
Case 16 |
|
|
SP |
NEP |
SP |
NEP |
SP |
NEP |
Pathogen identification
|
E.coli |
E.coli |
E.coli |
E.coli |
E.coli |
E.coli |
Amikacin |
S |
S |
S |
S |
S |
S |
Ampicillin |
S |
S |
R |
R |
S |
S |
Ampicillin/ Sulbactam |
S |
S |
R |
R |
S |
S |
Aztreonam |
S |
S |
R |
R |
S |
S |
Nitrofurantoin |
|
|
I |
I |
S |
S |
Trimethoprim/ sulfa |
S |
S |
R |
R |
S |
S |
Ciprofloxacin |
S |
S |
R |
R |
S |
S |
Meropenem |
S |
S |
S |
S |
S |
S |
Piperacillin |
S |
S |
R |
R |
S |
S |
Piperacillin/ Tazobactam |
S |
S |
|
|
S |
I |
Gentamicin |
S |
S |
R |
R |
S |
S |
Cefepime |
S |
S |
S |
S |
S |
S |
Cefuroxime |
S |
S |
R |
R |
S |
S |
Cefuroxime Axetil |
S |
S |
R |
R |
S |
S |
Ceftriaxone |
S |
S |
R |
R |
S |
S |
Ceftazidime |
S |
S |
R |
R |
S |
S |
Cefotetan |
S |
S |
S |
S |
S |
S |
Tobramycin |
S |
S |
R |
R |
S |
S |
Imipenem |
S |
S |
S |
S |
S |
S |
Levofloxacin |
S |
S |
R |
R |
S |
S |
E.coli: Escherichia coli; S: Sensitive; R: Resistant; I: Intermediate; *The kappa consistency Test was used, Kappa value = 0.955, 95% CI: 0.852-1.000, Kappa > 0.9, indicating that the consistency was excellent.
Table 4. Antibiotics susceptibility tests for Acinetobacter baumannii with two procedures*
Case 17 |
SP |
NEP |
Pathogen identification
|
Acinetobacter baumannii |
Acinetobacter baumannii |
Gentamicin |
S |
S |
Levofloxacin |
S |
S |
Aztreonam |
I |
I |
Piperacillin |
S |
S |
Piperacillin/ |
S |
S |
Tazobactam |
|
|
Cefepime |
S |
S |
Ceftriaxone |
I |
I |
Ceftazidime |
S |
S |
Cefotetan |
S |
S |
Tobramycin |
S |
S |
S: Sensitive; R: Resistant; I: Intermediate; *The kappa consistency test was used, Kappa value = 1.000, indicating that the consistency of SP and NEP was excellent.
Table 5. Antibiotics susceptibility tests for Enterococcus faecium with two procedures*
Case 18 |
SP |
NEP |
Pathogen identification |
Enterococcus faecium |
Enterococcus faecium
|
Nitrofurantoin |
R |
R |
Erythroprim/sulfa |
R |
R |
Ciprofloxacin |
R |
R |
Clindamycin |
R |
R |
Quinupristin/dalfopristin |
S |
S |
Moxifloxacin |
R |
R |
Tetracycline |
S |
S |
Vancomycin |
S |
S |
Levofloxacin |
R |
R |
Benzylpenicillin |
R |
R |
Linezolid |
S |
S |
Ampicillin |
R |
R |
S: Sensitive; R: Resistant; *The kappa consistency test was used, Kappa value =1.000, indicating that the consistency of SP and NEP was excellent.
Table 6. Antibiotics susceptibility tests for Pseudomonas aeruginosa with two procedures*
Case 19 |
SP |
NEP |
Pathogen identification |
Pseudomonas aeruginosa |
Pseudomonas aeruginosa
|
ciprofloxacin |
S |
S |
Levofloxacin |
S |
S |
amikacin |
S |
S |
meropenem |
S |
S |
Piperacillin |
S |
S |
Piperacillin/ tazobactam |
S |
S |
gentamicin |
S |
S |
cefepime |
S |
S |
Ceftazidime |
R |
S |
Tobramycin |
S |
S |
Imipenem |
S |
S |
S: Sensitive; R: Resistant; *The kappa consistency test was used, Kappa value > 0.9, indicating that the consistency of SP and NEP was excellent.
Table 7. Antibiotics susceptibility tests for Enterobacter aerogenes with two procedures*
Case 20 |
SP |
NEP |
Pathogen identification |
Enterobacter aerogenes |
Enterobacter aerogenes
|
Amikacin |
S |
S |
Ampicillin/ sulbactam |
R |
R |
Nitrofurantoin |
I |
I |
Trimethoprim/ sulfa |
R |
R |
Ciprofloxacin |
S |
S |
Meropenem |
S |
S |
Piperacillin |
R |
R |
Piperacillin/ tazobactam |
I |
S |
Gentamicin |
S |
S |
Cefepime |
S |
S |
Cefuroxime |
R |
R |
Ceftriaxone |
R |
R |
Ceftazidime |
R |
R |
Tobramycin |
S |
S |
Imipenem |
I |
S |
Levofloxacin |
S |
S |
Cefazolin |
R |
R |
S: Sensitive; R: Resistant; I: Intermediate; *The kappa consistency test was used, Kappa value =0.805, 95% CI: 0.564-1.000, Kappa > 0.9, indicating that the consistency of SP and NEP was excellent.
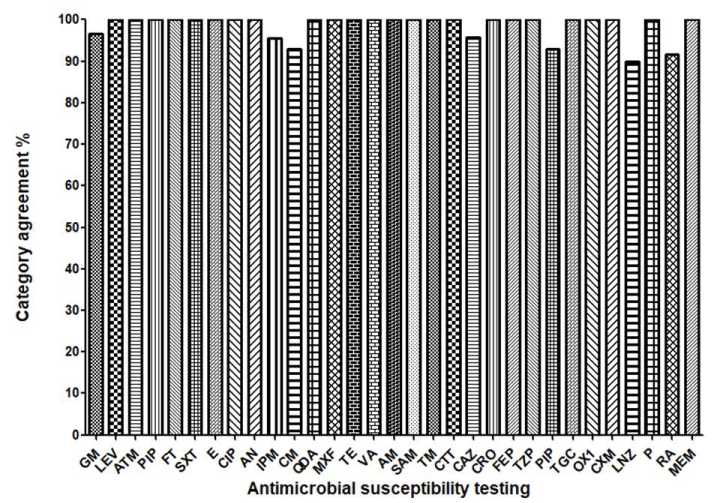
Figure 2C. The antibiotic sensitivity agreement between SP and NEP: A panel of antibiotics indicated in VITEK 2AST-GP67 and VITEK 2AST-GN09 cards were used in AST , including Gentamicin(GM), Levofloxacin(LEV), Aztreonam (ATM), Piperacillin(PIP), Nitrofurantoin(FT),Trimethoprim/sulfa(SXT), Erythroprim/sulfa(E), Ciprofloxacin (CIP), Amikacin (AN), Imipenem(IPM),Clindamycin(CM), Quinupristin/dalfopristin(QDA), Moxifloxacin(MXF), Tetracycline(TE), Vancomycin(VA), Ampicillin (AM), Ampicillin-sulbactam(SAM),Tobramycin(TM), Cefotetan (CTT), Ceftazidime (CAZ), Ceftriaxone (CRO), Aztreonam cefepime (FEP), Piperacillin(TZP), Piperacillin (PIP), Tigecycline(TGC), Oxacillin(OX1),Cefuroxime (CXM), Linezolid(LNZ), Penicillin-G/ Benzylpenicillin(P), Rifampin (RA), Meropenem ( MEM), etc. The AST results of SP and NEP had a 97.38% agreement rate. 4 (1.31%) minor errors were with linezolid (LNZ), Piperacillin (PIP) and Imipenem (IPM), respectively; and 4 (1.31%) major errors were with clindamycin (CM), Rifampin (RA), Gentamicin (GM) and Ceftazidime (CAZ), respectively(the kappa consistency test, p > 0.9).
In this study, we proved that our NEP has the following advantages: (1) shortening for about one day in the turnaround time of PI and AST, which is critical for appropriate treatment and rescue the life of patients with sepsis; (2) compared with the SP, our NEP yielded identical result in PI and 97.38% agreement in AST, indicating that NEP is acceptable without lowing the laboratory test standard; (3) omitted one step of the inoculation of bacteria from bottle to dish, which not only advancing the whole process for about one day, but also help in saving lab manpower and materials, which is benefit to laboratory staff and patients; (4) due to only omitted one step without changing any other process and devices/reagents used, it should be easy to apply to hospital using their currently used device, such as the VITEK 2 system (bioMerieux Inc) for PI and AST.
Again, this simple omitted one step modification is based on the facts that: (A) in most of cases, the invaded pathogen that causes sepsis is only single strain; (B) the SP does not separate colonies but collects all colonies from overnight cultured-dish, which allows us to omit this re-culture step and directly take 10 ml positive media to enrich the pathogen for PI and AST. Of cause, we do not against the day 2 dish culture for other purposes; such as further explore the other biological properties of pathogen. Since there are at least 20 ml media plus blood in the BD BACTEC bottle, taken 10 ml to perform an early diagnosis for the urgent needs of clinical medical decision is practicable and should be of benefit to patients.
Indeed, Accelerate Pheno™ System and Accelerate PhenoTest™ BC Kit, a newly FDA cleared fast diagnostic testing system (Accelerate Diagnostics Inc, 3950 S. Country Club Road, Tucson, Arizona, USA 85714) uses the same concept of one strain invaded to cause sepsis and directly takes microbes in positive blood culture bottle for PI and AST. As claimed by Accelerate Diagnostics Inc, their fully automated new device for AST could be less work, less workflow, at least 40h fast to yield report, less patient wait and shorter hospital stay (http://acceleratediagnostics.com). The use of Accelerate Pheno™ System provides a promising data in clinical setting [17,19]. The results of AST of direct use of broth from positive blood cultures and inoculated re-culture dish were also compared among the BacT/ALERT, BACTEC and VersaTREK systems, and the categorical agreement analysis was also very promising [18]. Increasing efforts are focused on direct use of broth of positive culture instead of re-inoculation to dish to obtain the isolates. Along this line, while several automation devices have been developed [19-20], the price of devices is relatively high, so that most of the hospitals or laboratories are reluctant to use the new devices and reagents with 1-2 delaying PI and AST reports [12]. Our NEP is an easy and economical alternative approach to achieve at least one day earlier report as newly developed devices without changing currently used device and reagents. By omitted one step, NEP is more cost-effective. Once the automation devices become cheaper and yield more advantages with FDA clearance, then, it can be shifted to use the advanced devices and reagents.
The bottle neck for this NEP is that the 10 ml culture media should be cleaned up both blood and other media portion as much as possible for not to interfere the following PI and AST assays. Several cautions need to be taken: (1) completely lyse the blood cells, since the bio-properties of cell plasma membrane and bacterial wall are quite different, the lysis time can be up to 12-18 min to ensure the blood cells are all lysed; (2) use sterilized saline or water to spin –washout the bacterial pellets; (3) collect only white pellets mainly with bacterial and justify to 0.5 McF to fit the assay needs; and (4) avoid any contamination in reagents and materials used during the operation process.
Although it is a simple modification of omitted one step without changing current device and reagent setting in any hospital, it really yields a great benefit of rescuing the sepsis patients by giving one day in advance for physicians to make a right antibiotics selection.
With the emergence of antibiotic-resistant bacteria as an increasing cause of sepsis death [21-22], we need to pay more attention not only in the fast reporting AST, but also the assays that detect the antibiotic-resistant bacteria. More studies need to be carried out to confirm the NEP efficacy and develop new assays for antibiotic-resistant bacteria.
This study was supported in part by grants from: International Science & Technology Cooperation Program of China (2015DFA31770) to JL; Fujian Development and Reform Commission to JL (FGW2014). We thank Dr. Shimin Zhang for editing this manuscript for publication.
Authors declare no duality of interest.
- Schuts EC, Hulscher MEJL, Mouton JW, Verduin CM, Stuart JWTC, et al. (2016) Current evidence on hospital antimicrobial stewardship objectives: a systematic review and meta-analysis. Lancet Infect Dis 16: 847-856. [Crossref]
- Pfafflin A, Schleicher E (2009) Inflammation markers in point-of-care testing (POCT). Anal Bioanal Chem 297: 1473-1480. [Crossref]
- Iregui M, Ward S, Sherman G, Fraser VJ, Kollef MH (2002) Clinical importance of delays in the initiation of appropriate antibiotic treatment for ventilator-associated pneumonia. Chest 122: 262-268. [Crossref]
- Luna CM, Aruj P, Niederman MS, Garzón J, Violi D, et al. (2006) Appropriateness and delay to initiate therapy in ventilator-associated pneumonia. Eur Respir J 27:158-164. [Crossref]
- Kibe S, Adams K, Barlow G (2011) Diagnostic and prognostic biomarkers of sepsis in critical care. J Antimicrob Chemother 66 Suppl 2: ii33-40. [Crossref]
- Barenfanger J, Drake C, Kacich G (1999) Clinical and financial benefits of rapid bacterial identification and antimicrobial susceptibility testing. J Clin Microbiol 37: 1415-1418. [Crossref]
- Doern GV, Vautour R, Gaudet M, Levy B (1994) Clinical impact of rapid in vitro susceptibility testing and bacterial identification. J Clin Microbiol 32: 1757-1762. [Crossref]
- Turner KM, Christensen H, Adams EJ, McAdams D, Fifer H, et al. (2017) Analysis of the potential for point-of-care test to enable individualised treatment of infections caused by antimicrobial-resistant and susceptible strains of Neisseria gonorrhoeae: a modelling study. BMJ Open 6: e015447. [Crossref]
- Beuving J, van der Donk CF, Linssen CF, Wolffs PF, Verbon A (2011) Evaluation of direct inoculation of the BD PHOENIX system from positive BACTEC blood cultures for both Gram-positive cocci and Gram-negative rods. BMC Microbiol 30: 156. [Crossref]
- Granato PA(1993)The impact of same-day tests versus traditional overnight testing. Diagn Microbiol Infect Dis 16: 237-243. [Crossref]
- Chandrasekaran S, Abbott A, Campeau S, Zimmer BL, Weinstein M, et al. (2018) Direct-from-blood culture disk diffusion to determine antimicrobial susceptibility of Gram-negative bacteria: Preliminary report from the Clinical and Laboratory Standards Institute Methods Development and Standardization Working Group. J Clin Microbiol pii: JCM.01678-17. [Crossref]
- Tabak YP, Vankeepuram L, Ye G, Jeffers K, Gupta V, et al. (2018) Blood Culture Turnaround Time in US Acute Care Hospitals and Implications for Laboratory Process Optimization. J Clin Microbiol pii: JCM.00500-18. [Crossref]
- Pulido MR, García-Quintanilla M, Martín-Peña R, Cisneros JM, McConnell MJ (2013) Progress on the development of rapid methods for antimicrobial susceptibility testing. J Antimicrob Chemother 68: 2710-2717. [Crossref]
- Murray MT, Johnson CL, Cohen B, Jackson O, Jones LK, et al. (2018) Use of antibiotics in paediatric long-term care facilities. J Hosp Infec 99: 139-144. [Crossref]
- Bauer M, Reinhart K (2010) Molecular diagnostics of sepsis--where are we today? Int J Med Microbiol 300: 411-413. [Crossref]
- American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference (1992) Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Crit care med 20: 864-874. [Crossref]
- Greenhalgh DG, Saffle JR, Holmes JH 4th, Gamelli RL, Palmieri TL, et al. (2007) American Burn Association consensus conference to define sepsis and infection in burns. J Burn Care Res 28: 776-790. [Crossref]
- Hanisch E, Brause R, Paetz J, Arlt B (2011) Review of a large clinical series: Predicting death for patients with abdominal septic shock. J Intensive Care Med 26: 27-33. [Crossref]
- Marschal M, Bachmaier J, Autenrieth I, Oberhettinger P, Willmann M, Peter S (2017) Evaluation of the Accelerate Pheno System for Fast Identification and Antimicrobial Susceptibility Testing from Positive Blood Cultures in Bloodstream Infections Caused by Gram-Negative Pathogens. J Clin Microbiol 55: 2116-2126. [Crossref]
- Chandrasekaran S, Abbott A, Campeau S, Zimmer BL, Weinstein M, et al. (2018) Direct-from-blood culture disk diffusion to determine antimicrobial susceptibility of Gram-negative bacteria: Preliminary report from the Clinical and Laboratory Standards Institute Methods Development and Standardization Working Group. J Clin Microbiol pii: JCM.01678-17. [Crossref]
- Ersoy SC, Heithoff DM, Barnes L, Tripp GK, House JK, et al. (2017) Correcting a Fundamental Flaw in the Paradigm for Antimicrobial Susceptibility Testing. EBioMedicine 20: 173-181. [Crossref]
- Kaushik AM, Hsieh K, Chen L, Shin DJ, Liao JC, et al. (2017) Accelerating bacterial growth detection and antimicrobial susceptibility assessment in integrated picoliter droplet platform. Biosens Bioelectron 97: 260-266. [Crossref]




