Oral squamous cell carcinoma of the gingivobuccal region (OSCC–GB) is the most prevalent cancer among males and fifth most prevalent among females from the Indian subcontinent [1]. Of about 350,000 new cases that arise worldwide each year; 119,992 new cases are contributed by India in 2018 [2]. We have previously identified the nuclear driver genes [3], important structural variations in chromosomes [4] and potentially druggable pathways [5] in OSCC-GB. We have also reported somatic driver mutations in mtDNA harboured by oral cancer patients [6,7]. In this report, we provide information which might be useful in deciphering the interplay between the nDNA and the mtDNA in tumourigenesis. In order to understand this, we have studied the correlation between somatic mtDNA mutations and somatic mutations in the nuclear driver genes from 89 gingivobuccal oral cancer patients.
Patients of OSCC-GB were recruited with informed consent [3]. Mitochondrial and nuclear DNA mutations were identified using massively parallel sequencing (NGS) data from paired DNA samples from tumour and matched blood from 89 patients. Sample BAM files used in this study are deposited in the European Genome Phenome Archive under accession code EGAS00001001901 (WGS), EGAS00001000249 & EGAS00001001028 (WES) and EGAD00001004987 (mtDNA sequence).
Ten nuclear genes (TP53, FAT1, CASP8, USP9X, MLL4, NOTCH1, HRAS, UNC13C, ARID2 and TRPM3) frequently mutated in OSCC-GB have been reported previously [3]. Among the 89 patients, 55 patients (61%) harboured somatic mutations in TP53. FAT1 or CASP8 were mutated in 26 patients each, followed by NOTCH1 (19 patients). Mutations in the mitochondrial genome in these patients were categorized into 17 features which are as follows; 13 mitochondrial coding genes as 13 individual features, 2 rRNAs (12S and 16S) and the D-loop region each as one feature and all mitochondrial tRNAs as a single feature. We found that mutations in ARID2 positively correlated with mutations in D-loop, mt-tRNA and ND4.
Somatic mutations in four nuclear driver genes, CASP8, NOTCH1, USP9X and TRPM3 displayed positive correlation with mutations in mitochondrial genes COX3, ND5, ND1 and COX2 respectively. Mutations in TP53 were found to be negatively correlated with somatic mutations in mitochondrial 12S rRNA and COX3 gene. Additionally, we observed that TP53 mutations were negatively correlated with mutations in NOTCH1, HRAS and USP9X. Thus, NOTCH1 and USP9X simultaneously appeared to be negatively correlated with TP53 mutations and positively correlated with some mitochondrial gene mutations (Figure 1A). Mutations in rest of the driver genes, namely FAT1, HRAS, UNC13C and MLL4 were not significantly correlated with somatic mtDNA mutations.
We also investigated the relationship between the mutation load of nuclear driver genes and mutation load of mtDNA in our dataset. We have categorized mutation burden of nuclear driver genes in each patient in 3 groups; mutation burden in all driver genes, mutation burden in TP53 and mutation burden in 9 other non-TP53 driver genes. Similarly, total somatic mutation burden in mtDNA was categorized into 4 groups; cumulative mutation burden in mitochondrial protein coding genes, mutation burden in D-loop as this is the control region for transcription and translation of mtDNA, mutation burden in all non- coding regions which includes D-loop, 2 mitochondrial rRNAs and 22 mitochondrial tRNAs, and total mutation burden which combines all the categories mentioned (Figure 1B). Spearman correlation test was performed between each of the nuclear mutation groups and individual mtDNA mutation groups. The mtDNA coding mutation load and mutation burden in 9 non TP53 driver genes was found to be significantly positively correlated (ρ = 0.2700105 P = 0.02087).
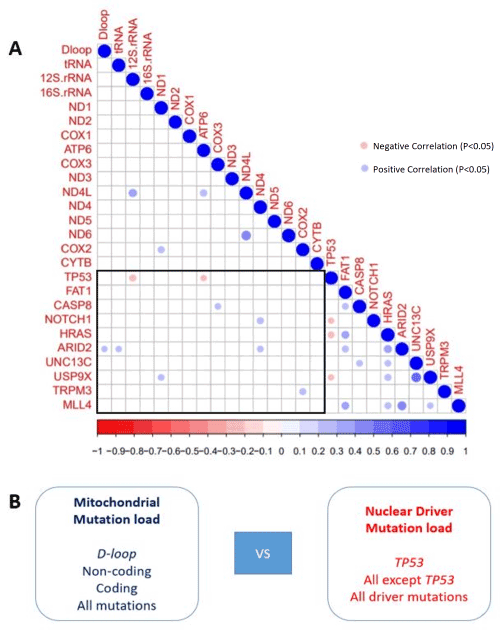
Figure 1: (A) Correlation between mitochondrial and nuclear mutations: Red circles indicate negative correlation and blue circles indicate positive correlation. Only significantly correlated genes are reported. (B) Schematic diagram representing the correlated mutation burden groups: Mutation loads in mitochondrial D-loop region, non-coding genes, coding genes and all of these mutations combined were compared with mutations load in nuclear driver genes, TP53 gene and all genes except TP53 to analyse mito-nuclear interaction at genome level.
In this study, we have also investigated the co-expression of 1701 nuclear coded genes which are associated with structure and function of mitochondria and mtDNA coded respiratory complex genes. This analysis did not throw any light on the retrograde signalling pathway which might be associated with oral cancer (result not shown). This might be attributed to post-transcriptional regulation of mitochondrial RNA processing, which promote the difference in abundance of 13 mitochondrial transcripts [6,7], despite that they are transcribed as a single poly-cistronic RNA [8]. Also translational modulation of nuclear and mitochondrial genes regulate the mitochondrial and nuclear OXPHOS protein levels which might be necessary for the various subunits of the electron transport chain to assemble at a specific stoichiometric ratio [9]. The above reports along with our observations point towards the possible shortcoming of gene expression data as surrogate for mito-nuclear interaction at RNA level. Although altered expression of mitochondrial and nuclear OXPHOS genes independently, might indicate potential reprograming of the tumour metabolism [10].
We have observed that patients harbouring mutations in other nuclear driver genes such as HRAS, NOTCH1, USP9X etc., are more likely to harbour mutations in mitochondrial genes. We also found that most of the patients carrying mutations in TP53 are less likely to harbour mutations in mtDNA. These observations suggest that in the absence of mutations in TP53, mutations in mtDNA and other driver genes might aid in the progression of cancer in a complementary fashion. The possible mechanism via which mtDNA coding mutations and mutations in 9 non-TP53 driver genes aid in tumour progression in absence of TP53 loss of function mutations needs further investigation. A version of this manuscript has been deposited at bioRxiv (doi: https://doi.org/10.1101/714519).
- National Institute of Cancer Prevention and Research. [http://cancerindia.org.in/oral-cancer/] (Accessed on 26 April 2019).
- Globocan 2018. []http://gco.iarc.fr (accessed on 26 April 2019).
- India Project Team of the International Cancer Genome Consortium (2013) Mutational landscape of gingivo-buccal oral squamous cell carcinoma reveals new recurrently-mutated genes and molecular subgroups. Nat Commun 4: 2873. [Crossref]
- Biswas NK, Das C, Das S, Maitra A, Nair S, et al. (2019) Lymph node metastasis in oral cancer is strongly associated with chromosomal instability and DNA repair defects. Int J Cancer 145: 2568-2579. [Crossref]
- Biswas NK, Das S, Maitra A, Sarin R, Majumder PP (2014) Somatic mutations in arachidonic acid metabolism pathway genes enhance oral cancer post-treatment disease-free survival. Nat Commun 5: 5835. [Crossref]
- Rorbach J, Minczuk M (2012) The post-transcriptional life of mammalian mitochondrial RNA. Biochem J 444: 357-373. [Crossref]
- Palodhi A, Ghosh S, Biswas NK, Basu A, Majumder PP, et al. (2018) Profiling of genomic alterations of mitochondrial DNA in gingivobuccal oral squamous cell carcinoma: Implications for disease progress. Mitochondrion 46: 361-369. [Crossref]
- Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, et al. (2011) The human mitochondrial transcriptome. Cell 146: 645-658. [Crossref]
- Couvillion MT, Soto IC, Shipkovenska G, Churchman LS (2016) Synchronized mitochondrial and cytosolic translation programs. Nature 533: 499-503. [Crossref]
- Reznik E, Wang Q, La K, Schultz N, Sander C (2017) Mitochondrial respiratory gene expression is suppressed in many cancers. Elife 6: e21592. [Crossref]

