Breast cancer is the world leading type of cancer for women, 2 million cases in 2018 [1]. Epidermal growth factor receptor (EGFR) has been associated with cancer cell transformation, marking it as an attractive target for therapeutic strategies. Its overexpression in the early stages of breast cancer is associated with reduced chances of malignancy remission and overall survival [2].
Nowadays, for breast cancer patients, measuring EGFR requires complex instrumentation which necessitates clinically unrealistic time, expense and personnel. It is then a necessity to develop a more practical method that is sensitive, simple, fast, and low-cost for point-of-care detection of EGFR [3].
Biosensors are devices that turn biological interaction into a physical signal. This interaction gets registered on a transducer (electrochemical, optical, mechanical…) that then turns it into an electronic signal [4-6].
Screen-Printed electrodes (SPE) technology offers a promising approach toward that solution as they fulfill most of the requirement mentioned above and, that is why SPE have high flexibility for a variety of commercial uses [7].
In this paper we present the preliminary results on the development of an electrochemical gold integrated SPE biosensor for the detection of EGFR. The strategy followed concerned the biofunctionalization of the gold electrode through the in-situ electrochemical deposition of 4-carboxymethylanailine (CMA) then the grafting of Anti-EGFR antibodoy. The biosensor developed was characterized using electrochemical impedance spectroscopy (EIS) and as a result it was found to be highly sensitive within a range of 50-120 pg/mL in phosphate buffer saline (PBS) as well as very selective toward the EGFR when compared to other interferences such as HER2 and HER3.
Chemical products
The reagents used in this research are: 4-aminophenylacetic acid (4-carboxymethylaniline (CMA)), sodium nitrite (NaNO2), hydrochloric acid (HCl), N-hydroxysuccinimide (NHS), N-(3-dimethylaminopropyl)-N-ethyl-carbodiimide hydrochloride(EDC), ethanolamine, PBS, phosphate buffered saline Tween (PBS-Tween) and (Fe2+Fe3+) were purchased from Sigma Aldrich (France). EGFR protein and anti-EGFR antibody were purchased from R&D System (France).
Antibodies (Ab) and antigen (Ag) solutions
As per the supplier’s protocol, the antibodies and the antigens have been diluted in PBS (pH 7.4) to acquire a stock solution with a final concentration of 10 µg/mL. for the antigen (EGFR) and 0.5 mg/mL for the antibody (anti-EGFR), which were then divided into aliquots and stored at -20°C. The stock solutions for EGFR was then further diluted in PBS to form the standard solutions used in the analysis (50, 80, 100, 120 pg/mL).
Electrochemical characterization
All electrochemical measurements were carried out at room temperature (20 ± 2°C) in a Faraday box using a PalmSense 4 potentiostat (Palmsense B.V., Netherlands) controlled by PSTrace software (version 5.0.5, Palmsense B.V., Netherlands). Cyclic Voltammetry (CV) was used for SPE characterization using the redox probe K3[Fe(CN)6]/K4[Fe(CN)6] at 5 mM in PBS buffer at pH7.4. The potential window that was used was between -0.4 and 0.6 V, and the scan rate is 0.06 V/s. CV was repeated three times until a stable measurement was achieved. Electrochemical impedance spectroscopy (EIS) measurements were used to evaluate the recognition properties of the immunosensing platform, for both sensitivity and selectivity. During the measurements, the potential was kept at 0.174 V versus the integrated Ag/AgCl reference electrode after the SPE was submerged in an electron mediator solution of 5 mM of K3[Fe(CN)6]/K4[Fe(CN)6] in PBS buffer at pH=7.4. Data acquisition and analysis were accomplished using the open access software EIS spectrum analyzer.
SPE activation using 4-carboxymethyl aniline
CV was used for 4-carboxymethyl aniline (CMA) electrochemical deposition, as described in [8-13]. The 3 mM CMA solution was prepared in deionized water through 10 min of sonication, then two solutions of HCl 1M and NaNO2 1M were added to form a concentration of 15 mM each. The solution was kept in the fridge for 10 min and then was mixed with the previously prepared CMA solution to create the diazoted derivative. 15 CV cycles in a window of -0.2 to -1.2 V at a scan rate of 20 mV/s were applied to the SPE in the CMA solution, enough to cover the entire SPE surface with CMA. The cyclic voltammogram of the electrodeposition can be seen in figure 1C.
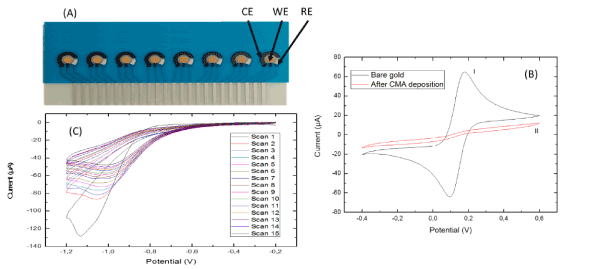
Figure 1: (A) biosensor platform containing 8 integrated SPE with each SPE containing a silver/silver chloride reference electrode (RE) a carbon paste counter electrode (CE) and a gold working electrode (WE). (B) Cyclic voltammogram of a WE before (I) and after (II) CMA electrochemical deposition. (C) the cyclic voltammogram for the electrodeposition of CMA on the WE surface
Ab immobilization on the SPE
Anti-EGFR was then immobilized on the CMA-modified SPE by first activating the carboxylic acid groups of CMA molecules through incubating them in EDC (0.4M)/NHS (0.1M) in deionized water for 40 min. Afterwards the device was washed with PBS to eliminate excess EDC/NHS, and immediately incubated with the antibody anti-EGFR in PBS (10 µg/mL) for 40 min at room temperature. The remaining carboxylic groups were blocked by incubating the SPE in ethanolamine solution (1‰ in PBS) at room temperature for 30 min to prevent nonspecific binding at the detection stage.
Bio-functionalization of the gold surface
The fully integrated biosensor platform used for EGFR detection was previously fabricated and characterized for many complex mediums (Figure 1A). The SPE is characterized by CV before and after CMA deposition as described in section 2.3. The results of this characterization are shown in figure 1B. The disappearance of the oxide/reduction peaks from bare gold to after deposition of CMA was caused by blocking of the CMA layer of the gold surface thus creating a weak electron kinetic transfer.
Detection and interferences of EGFR in PBS
A Nyquist plot (Np) presentation of the biosensor at different concentration of EGFR is shown in figure 2A. The largest semicircle corresponds to the immobilization of anti-EGFR. After incubation of the bio-functionalized WE in 50 pg/mL of EGFR, a second Np with a smaller semi-circle is obtained, confirming a change on the global electrochemical impedance of the sensor due to a specific binding. The Nps continued to decrease with the increase concentration of EGFR, indicating a good response toward the target analyte from the biosensor. The Nyquist plots were then fitted using Randles equivalent circuit model (Figure 2A), where the WE is presented as resistance (R1), the charge-transfer resistance (R2), the Warburg impedance (W1) and a constant phase element (CPE).
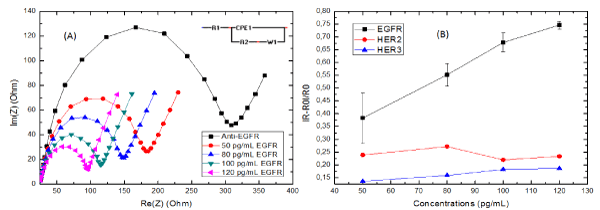
Figure 2: (A) Nyquist impedance plot (Re(Z) vs Im(Z) of K3[Fe(CN)6]/ K4[Fe(CN)6] (5mM) in PBS (pH=7.4) at various concentration of EGFR. (B) Detection curve of the biosensor bio-functionalized for the detection of EGFR, HER2 and HER3
The real impedance (Re(Z)) is represented as the x-axis in a Nyquist plot. It is the mathematical representation of R1 and R2 on that plot. So a decrease in the Nyquist plot size means a decrease with the R2 of the WE. With that decrease being related to the concentration of EGFR as observed in figure 2A, this means that a decrease of R2 is directly linked to the biosensor’s sensitivity. The normalized data were presented in fig 2(B) as |ΔR2|/R2 (Whereas |ΔR2|/R2 = |R2(EGFR)-R2(anti-EGFR)| / R2(anti-EGFR)) with an equation [Y = (4.77×10^-3 ± 4.74×10^-4) X + (0.1760 ± 0,0540)] with an R-square = 0.9709.
In order to test the biosensor’s selectivity, additional tests were carried out using the same bio-functionalization steps as mentioned before only with the detection of biomarkers other than EGFR. For that purpose, HER2 and HER3 were chosen. These biomarkers have a similar structure to EGFR [14,15], while several studies have found the presence of HER2 in saliva [16,17] it remains unclear whether HER3 is also present there. The sensitivity of the biosensor toward EGFR is higher in comparison to that of HER2 (Y = (-1.43×10^-4 ± 0,0011) X + (0.3946 ± 0.1116), R-square = -0.4934) and HER3 (Y = (7.94×10^-4 ± 9.8350×10^-5) X + (0.0963 ± 0.00803), R-square = 0.9554), proving its selectivity toward EGFR over the other two biomarkers.
In conclusion, we would like to report the successful electrodeposition of a passivation layer of CMA on a SPE as well as the successful immobilization of anti-EGFR antibody on that electrode. Preliminary tests showed a linear progression from 50 pg/mL to 120 pg/mL between EGFR concentration and the charge-transfer resistance of the working electrode (R-Square = 0.9709), as well as a minimal level of interferences attributed to nonspecific binding showing a good sensitivity toward the target biomarker (slope = 4.77×10^-3 ± 4.74×10^-4) when compared to the sensitivity toward other biomarkers (ex. HER2 (slope = -1.43×10^-4 ± 0,0011) and HER3 (slope = 7.94×10^-4 ± 9.8350×10^-5)). As such, this biosensor presents a good platform for future development of a tool for the detection of EGFR in saliva.
This research was supported by UK Lebanon techhub.
- Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, et al. (2018) Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer: 1-13. [Crossref]
- Gonzalez-Conchas GA, Rodriguez-Romo L, Hernandez-Barajas D, Gonzalez-Guerrero JF, Rodriguez-Fernandez IA, et al. (2018) Epidermal growth factor receptor overexpression and outcomes in early breast cancer: A systematic review and a meta-analysis. Cancer Treat Rev 62: 1-8. [Crossref]
- Ilkhani H, Sarparast M, Noori A, Zahra Bathaie S, Mousavi MF (2015) Electrochemical aptamer/antibody based sandwich immunosensor for the detection of EGFR, a cancer biomarker, using gold nanoparticles as a signaling probe. Biosens Bioelectron 74: 491-497. [Crossref]
- Bahadir EB, Sezgintürk MK (2016) A review on impedimetric biosensors. Artif Cells Nanomedicine Biotechnol 44: 248-262. [Crossref]
- Jayanthi VSPKSA, Das AB, Saxena U (2017) Recent advances in biosensor development for the detection of cancer biomarkers. Biosens Bioelectron 91: 15-23. [Crossref]
- Karunakaran C, Rajkumar R, Bhargava K (2015) Introduction to Biosensors. Biosens Bioelectron 60: 1-68.
- Taleat Z, Khoshroo A, Mazloum-Ardakani M (2014) Screen-printed electrodes for biosensing: A review (2008-2013). Microchim Acta 181: 865-891.
- Mohamed AA, Salmi Z, Dahoumane SA, Mekki A, Carbonnier B, et al. (2015) Functionalization of nanomaterials with aryldiazonium salts. Adv Colloid Interface Sci 225: 16-36. [Crossref]
- Cao C, Zhang Y, Jiang C, Qi M, Liu G (2017) Advances on Aryldiazonium Salt Chemistry Based Interfacial Fabrication for Sensing Applications. ACS Appl Mater Interfaces 9: 5031-5049. [Crossref]
- Baraket A, Lee M, Zine N, Sigaud M, Bausells J, et al. (2017) A fully integrated electrochemical biosensor platform fabrication process for cytokines detection. Biosens Bioelectron 93: 170-175. doi:10.1016/j.bios.2016.09.023. [Crossref]
- Bellagambi FG, Baraket A, Longo A, Vatteroni M, Zine N, et al. (2017) Electrochemical biosensor platform for TNF-a cytokines detection in both artificial and human saliva: Heart failure, Sensors Actuators. B Chem 251: 1026-1033.
- Baraket A, Lee M, Zine N, Sigaud M, Yaakoubi N, et al. (2013) Diazonium modified gold microelectrodes onto polyimide substrates for impedimetric cytokine detection with an integrated Ag/AgCl reference electrode. Sensors Actuators B Chem 189: 165-172.
- Ghedir E, Baraket A, Kouchar S, Rabai S, Benounis M, et al. (2018) Electrochemical Immunosensor for NT-proBNP Detection in Artificial Human Saliva: Heart Failure Biomedical Application. Proc 2,
- Sierke SL, Cheng K, Kim HH, Koland JG (1997) Biochemical characterization of the protein tyrosine kinase homology domain of the ErbB3 (HER3) receptor protein. Biochem J 322: 757-763. [Crossref]
- Omenn GS, Guan Y, Menon R (2014) A new class of protein cancer biomarker candidates: Differentially expressed splice variants of ERBB2 (HER2/neu) and ERBB1 (EGFR) in breast cancer cell lines. J Proteomics 107: 103-112. [Crossref]
- Laidi F, Bouziane F, Lakhdar A, Khabouze S, Rhrab B, et al. (2014) Salivary expression of soluble HER2 in breast cancer patients with positive and negative HER2 status. Onco Targets Ther 7: 1285-1289. [Crossref]
- Streckfus CF, Arreola D, Edwards C, Bigler L (2012) Salivary protein profiles among her2/neu-receptor-positive and -negative breast cancer patients: Support for using salivary protein profiles for modeling breast cancer progression. J Oncol.


