Abstract
The intracellular solubility and folding properties of proteins are influenced by numerous extrinsic and intrinsic factors. One of the intrinsic parameters is the presence of an aggregating motifs within the amino acid sequence of the protein which makes the protein intrinsically disorder. Here we have studied the effect of an ultrashort self-assembling peptide (SSP) motif consisting of four amino acids, Isoleucine-Valine-Phenylalanine-Lysine, on the aggregation propensity of intracellular expression and folding of small ubiquitin-like modifier (SUMO) protein by c-terminally conjugation with the SSP motif. Conjugation of the SSP motif directed SUMO accumulation into the inclusion bodies of E. coli. SSP conjugated inclusion bodies of SUMO have revealed a highly ordered, rich fibrous structure, when observed under scanning electron microscopy. FTIR analysis of SSP conjugated SUMO has also confirmed that the composition of self-assembling fibrous material is mainly beta sheet rich. This observation has confirmed our hypothesis that SSP motifs can lead to the aggregation of soluble proteins, when exposed to favorable conditions.
Keywords
aggregation, self-assembling peptide, inclusion bodies, protein expression
Introduction
Proteins are vital for virtually all biological processes and, since native expression does not yield sufficient amounts for structural and functional characterization, recombinant protein production is indispensable. Moreover, protein stability and yield are important prerequisites for a wide range of applications including an increasing number of therapeutic applications such as monoclonal antibodies, enzymes, cytokines and hormones [1,2].
Expression of recombinant proteins is hampered due to their poor solubility in heterologous hosts. Intracellular misfolding and poor solubility leads to the formation of inclusion bodies in the submicron sized species of bacteria. [3] Sometimes inclusion bodies based protein expression is deliberate mainly to boost the recombinant protein production [4] or reduce the toxicity of target protein to the host [5]. Some protein-based drugs are also packaged in inclusion bodies [6]. However most of the time recombinantly produced proteins are required to be soluble and fully folded. For this purpose, several N-terminal fusion tags like Maltose Binding protein (MBP), trigger factor (TF), Thioredoxin (TRX), transcription termination/antitermination (NusA), Ubiquitin protein (UB), glutathione S-transferase (GST) and small ubiquitin-like modifier (SUMO) have been used to improve the solubility of protein in heterologous prokaryotic and eukaryotic hosts [7]. Use of SUMO fusion have been reported to increase protein yield, solubility, and purification in prokaryotes [7,8].
Protein based pharmaceuticals are one of the innovative pharmaceuticals that hold promise as a multibillion dollar industry in the health sector [9]. Protein aggregation, however, causes a major economic and technical burden on pharmaceutical and biotechnology industries. For instance, protein based pharmaceutical products face major challenges due to their aggregation over time, as proteins have an inherent tendency to aggregate [10]. Protein aggregation is a process during which misfolded proteins accumulate into insoluble agglomerates. In order to be functional, proteins should be fully folded. Therefore, if the protein of interest is to be used as a therapeutic product, it should have long term stability. There are many in vitro and in vivo factors affecting the stability of proteins in certain solutions as the interacting forces play important for the folding of proteins. These interactions are mainly non-covalent which include could enhanced hydrogen bonding, hydrophobic attractions and van der Waals forces among backbone and side-chain atoms, low steric clashes and minimizing high energy interactions between amino acids and other components in solution [11].
Several neurodegenerative diseases related to aggregation of intrinsically disordered proteins and most famous include Parkinson’s, Alzheimer's, Dementia with Lewy bodies and the transmissible spongiform encephalopathies, frontotemporal dementias etc. [12]. Typical common characteristics of Prion and amyloid diseases include ordered proteins aggregate formation mostly in the form of long fibrils [13]. Most of these proteins are intrinsically disordered and lack stable native structure which in turn allows them the flexibility to adopt many conformations and perform important physiological functions. Once example of such proteins is microtubule associated protein TAU which is involved in regulation and stabilization of microtubules but dysfunction of TAU can lead to oligomerization and fibril formation in the brain causing many of the diseases mentioned above [14]. Many extrinsic and intrinsic factors modulate the aggregation behavior of intrinsically disordered proteins. Osmolytes are the examples of extrinsic factors regulating the aggregation properties of intrinsically disordered proteins [15,16].
The intrinsic aggregation propensity of proteins is directly related to presence of aggregation prone regions which reduces the solubility of these proteins [17]. In silico Aggregation prediction methods which are widely used and often rely on the formation of intermolecular beta sheets [18]. Recent studies have revealed the presence of at least one fibril forming segment in about 1% of human proteome [19]. Fourteen aggregation prone motifs have been identified in human immunoglobulin G [20]. However complete understanding of the mechanism underlying protein aggregation is still under debate [21]. Herein we describe, the impact of the aggregation triggering ultrashort self-assembling peptide (SSP) motif, consisting of four amino acids Isoleucine-Valine-Phenylalanine-Lysine, derived and modified from microtubule associated protein, TAU, on the solubility of SUMO. Multiple copies of aggregation triggering peptides were attached to SUMO in different constructs and their effect on the aggregation of SUMO was studied.
Materials and methods
Construct designing and preparation
Single stranded DNA oligonucleotides coding for corresponding USPs were first optimized using genart Invitrogen online codon optimization software. These single stranded DNA fragments and their reverse complementary strands with forward primer having 5’ AGGT and 3’ TAAT overhang. Similarly the reverse primer has 5’ CTAGATTA overhang as shown in the table. These DNA fragments were synthesized from Sigma-Aldrich. Approximately 600pM of each oligonucleotide was phosphorylated using 5 units of T4 polynucleotide kinase and 1x T4 DNA ligase buffer at 37 °C for 45 minutes in a small Eppendorf tube. The reaction was stopped by heating the tube at 65 C for 20 minutes. After phosphorylation equimolar concentration of complementary oligonucleotides were annealed in buffer containing 40mM Tris HCl pH 8.0, 10mM MgCl2, 50mM NaCl. The mixture was first heated at 99 C and gradually cooled 50 °C. Annealed double stranded DNA fragments were then purified by 1.5% agarose gel electrophoresis and Qiagen gel extraction kit.
Vector Preparation: Current studies were carried out using pE-SUMO vector with kanamycin resistance from life sensors. pE-SUMO vector was restricted with type IIS restriction enzyme Bsa1 overnight at 37 °C. The restricted vector was then the phosphorylated using alkaline phosphatase to avoid self-ligation of the vector.
For ligation 75-100ng of pE-SUMO was mixed with 100x molar excess of double stranded DNA fragments. AS both vector and insert have complementary sticky ends, therefore T4 DNA ligase was used for ligation and the ligation reaction was incubated overnight at 16 °C. About 4ul of ligation reaction was transformed to E. coli DH5α cells and transformation was performed as per stranded protocol and six colonies were screened by miniprep for each construct to find successful clones. Positive clones were confirmed by DNA sequencing with T7 forward primer.
Expression of SUMO-SSP constructs
Expression of SSP1, SSP2, SSP3 and SSP5 as well as control constructs was carried out in E. coli BL21(DE3) cells. About 200ng of each plasmid were transformed into BL21(DE3) cells and next day 3-4 colonies from plate were inoculated in 50ml of LB (Kanamycin) cells were grown to visible turbidity. These primary cultures were then diluted 100 times in 2L flasks containing 1L of LB (Kanamycin) and the cells were grown to OD600 of 0.8 at 37 °C before being induced with 0.5M isopropyl thio‐β‐D‐galactoside (IPTG) at 25 C for 16 hours. Cells were then harvested by pelleting them at 6000g for 20minutes.
Inclusion body isolation and Characterization
For isolation of inclusion bodies, cell pellets containing expressed constructs were resuspended in 50mM Tris+ 100mM NaCl+EDTA-free 1x protease inhibitor, pH 8.0. Cells were then sonicated for 20 cycles of 10 seconds-on and 20 seconds-off on ice to break the cells. Sonicated solution of lysed cells was spun at 13000g for 40 minutes. Pelleted inclusion bodies were then washed twice with 50mM Tris+100mM NaCl+0.5% triton x-100, pH 8.0 and once with 50mM Tris+100mM NaCl, pH 8.0 to remove cell debris. For denaturation of inclusion bodies, these were resuspended in denaturation buffer containing 50mM Tris+8M urea, pH 8.0 at 2mg/ml concentration using tissue homogenizer and bath sonication. 50mM Tris+6M GnHCl, pH 8.0 was also used to denature inclusion bodies.
Refolding and Purification of SUMO-SSP peptides
Denatured inclusion bodies solution for each SUMO-SSP construct were filtered through 0.45um filter before being loaded on to His-trap (5ml) affinity column under denaturing condition in 50mM Tris+8M urea, pH 8.0 buffer using AktaStart from GE healthcare. Protein coding for each construct was eluted from the column in 50mM Tris+100mM NaCl+2M urea+ 500mM Imidazole pH 8.0 by gradient elution. Elution fractions were screened by SDS-PAGE gel electrophoresis.
Characterization of SUMO-SSP peptides by scanning electron microscopy
The morphology of SUMO-SSP was determined using an FEI Magellan XHR or Quanta 600 scanning electron microscope (SEM) with an accelerating voltage of 2-5kV. The SEM samples were prepared either by lyophilization of SUMO-SSP solution followed by coating of powdered sample on sticky carbon tape or by putting directly a drop of SUMO-SSP on a silicon wafer and drying in a vacuum desiccator for overnight. Lastly, the dried samples from both sample preparation methods were sputter coated with 5 nm Iridium prior to imaging.
Fourier-transform infrared spectroscopic analysis of SUMO-SSP fusion proteins
FTIR measurements of SUMO-SSP fusion proteins were taken by Thermo Scientific FTIR-ATR iS10. A background scan was measured before the sample. The spectrum was collected in the range 500-4000 cm-1, with a 1 cm-1 interval. Both background and sample measurements were taken as average over 10 scans
Results and discussion
Gene designing and vector construction
Codon optimized single stranded DNA fragments coding for SSP motifs were designed in such a way that the SSP motifs were directly conjugated to the SUMO protein sequence (Figure 1). For this purpose pE-SUMO was restricted with type IIS secretion enzyme Bsa1 in such a way that it created a four nucleotide overhang on lower strand (3’). Complementary four nucleotides were added to the 5’ end of all forward DNA fragments. The single stranded DNA sequences as described in the table 1, were synthesized from Sigma-Aldrich. All the lypholized DNA fragments were solubilized in autoclaved MiliQ H2O at a concentration of 100 μM. To facilitate the annealing, phosphorylation of all single stranded DNA fragments was carried out using T4 poly nucleotide kinase. These single stranded DNA fragments were then annealed together. As the DNA fragments coding for SSP motifs are small compared to the size of the vector pE-SUMO, therefore 100x molar access of these fragments was used in a ligation reaction. Ligation reaction once transformed to E. coli DH5α cells, produced colonies on LB agar plates containing kanamycin antibiotic. Since the pE-SUMO vector was dephosphorylated using alkaline phosphatase to avoid the self-ligation of the vector, therefore the colonies appeared on the kanamycin containing LB agar plates were predominately SSP motifs ligated vector. Further confirmation of these clones was confirmed by DNA sequencing of 4-6 plasmids harvested by miniprep. DNA sequencing of these plasmids confirmed the successful insertion of the DNA sequence coding for SSP motifs.
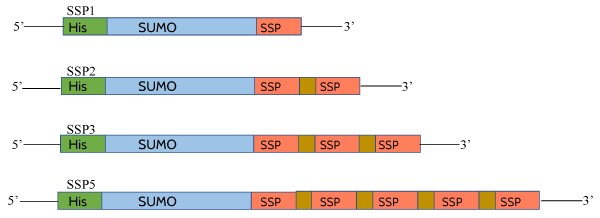
Figure 1. Template of sumo-conjugated SSPs. Separate constructs were made with different number of copies of SSPs. SSP1 contains one copy of motif, SSP2 with two copies of motif separated by GPG linker, similarly SSP3 and SSP5 are with three and five copies of motif. Each motif copy is separated by GPG linker.
Table 1. DNA sequences coding for SSPs. These sequences were synthesized from Sigma-Aldrich.
Name |
Oligonucleotide sequence |
SSP1_F |
AGGTATTGTTTTTAAATAAT |
SSP1_R |
CTAGATTATTTAAAAACAAT |
SSP2_F |
AGGTATTGTTTTTAAAGGTCCTGGCATCGTGTTTAAATAAT |
SSP2_R |
CTAGATTATTTAAACACGATGCCAGGACCTTTAAAAACAAT |
SSP3_F |
AGGTATTGTTTTTAAAGGTCCTGGCATCGTGTTTAAAGGCCCAGGTATTGTGTTCAAATAAT |
SSP3_R |
CTAGATTATTTGAACACAATACCTGGGCCTTTAAACACGATGCCAGGACCTTTAAAAACAAT |
SSP5_F |
AGGTGGTCCGGGTATTGTTTTTAAAGGTCCTGGCATCGTGTTTAAAGGCCCAGGTATTGTGTTCAAAGGACCGGGAATAGTATTCAAAGGCCCTGGGATTGTATTTAAAGGTCCCGGATAAT |
SSP5_R |
CTAGATTATCCGGGACCTTTAAATACAATCCCAGGGCCTTTGAATACTATTCCCGGTCCTTTGAACACAATACCTGGGCCTTTAAACACGATGCCAGGACCTTTAAAAACAATACCCGGACC |
Expression of SUMO-SSP constructs
Once confirmed by DNA sequencing, plasmids containing the SSP motifs were expressed in E. coli BL21(DE3) cells. As expected SUMO-SSP conjugated proteins expressed at higher levels when induced with IPTG, It was observed by western blotting that all of the SSP conjugated SUMO protein appeared in the inclusion bodies of E. coli, when blotted against anti-histidine antibodies. There was no band observed for soluble fraction which indicates that the presence of SSPs in the recombinant protein leads to its misfolding and the formation of inclusion bodies upon translation. Since the Histidine tag is on the N-terminus of SUMO protein, therefore it can be said that soluble SUMO protein once attached to the SSP motifs aggregates and forms inclusion bodies. This observation has confirmed our hypothesis that SSP motifs, when exposed to favorable conditions can lead to the aggregation of soluble proteins (Figure 2).
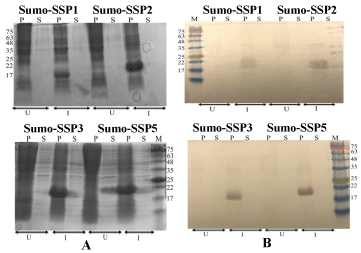
Figure 2. Expression of SUMO-SSPs constructs. Panel A SDS-PAGE followed by Coomassie staining Panel B SDS-PAGE followed by western blotting against anti-his antibodies. P stands for sample from pellet after cell lysis, S stand for supernatant after cell lysis. U stands for Un-induced samples, I stands for induction of samples with IPTG after cell density reach to OD600 0.8. M is pre-stained protein marker and prominent bands are indicated by their size in kDa.
Refolding and characterization of SUMO-SSP conjugated proteins
For further characterization of SUMO-conjugated SSP motifs, expression of these conjugated proteins was scaled up to 1L and inclusion bodies were prepared by cell disruption. Since most of the inclusion bodies contains small fraction of impurities, therefore the mild detergent solution was used to remove impurities. Intact inclusion bodies were similar to those reported in the literature when observed by SEM (Figure 3)[22-24].
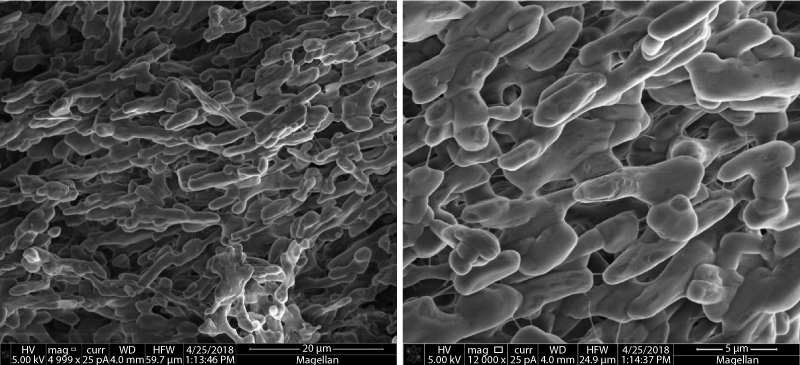
Figure 3. Scanning electron microscopy of inclusion bodies of SUMO-SSP5.
These inclusion bodies were then denatured using denaturants like GnHCl and Urea. It was observed that SUMO-conjugated SSP motifs were more soluble in urea compared to GnHCl. It was also noted that inclusion bodies tend to solubilize more in basic denaturant solution when compared to acidic solution. It was also found that inclusion bodies adhesiveness increased with an increasing number of SSP copies i.e. SUMO-SSP1<SUMO-SSP2<SUMO-SSP3<SUMO-SSP5. It means SUMO-SSP1 inclusion bodies were easily soluble and more stable in denaturant solution compared to SUMO-SSP5. Solution containing ammonium acetate and acetic acid was less effective compared to Tris and Urea. The denatured inclusion bodies for SUMO-conjugated SSP motifs, when observed under SEM shown similar structure (Figure 4).
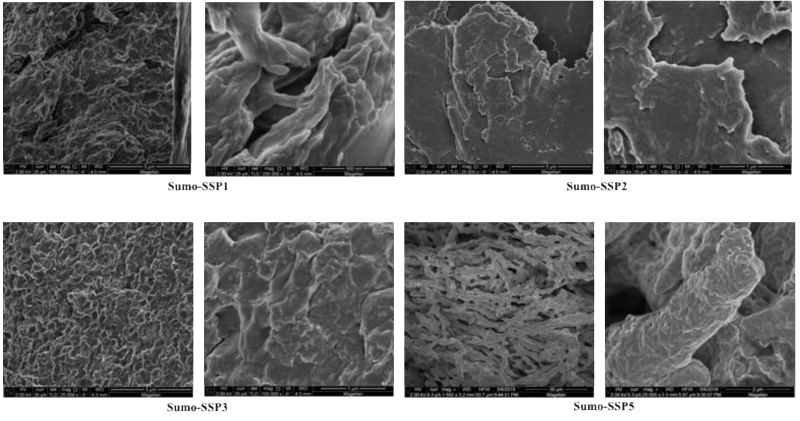
Figure 4. Morphology characterization of SUMO-SSPs inclusion bodies solubilization by scanning electron microscopy in the presence of denaturant 8M urea.
Refolding was initially attempted by dialyzing the denatured SUMO-SSP solution against buffers without denaturant at 4 °C, however it was observed that SUMO-SSP proteins tend to precipitate during overnight dialysis. Refolding was therefore carried out by nickel affinity chromatography. SUMO-SSP solutions were applied to the His-trap 5ml column under denaturing conditions and proteins were refolded and eluted from column in a buffer containing reduced urea concentration (1M) and imidazole. Eluted proteins when observed by SDS-PAGE gel electrophoresis, produced bands for purified SUMO-SSP conjugated proteins (Figure 5). His-trap affinity chromatography based purification results have shown that high quality purification and refolding of SUMO-SSPs can be performed in a single step. These refolded and purified proteins did not precipitate when dialyzed. This is indicative of the increased stability of the SUMO-SSP conjugated proteins after refolding.
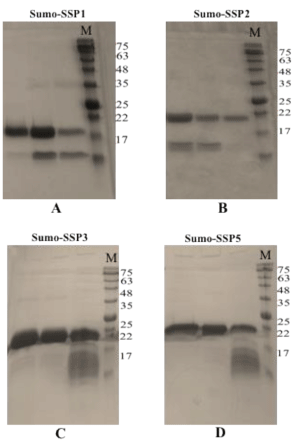
Figure 5. Purification of SUMO-SSPs. A SUMO-SSP1, B SUMO-SSP2, C SUMO-SSP3, D SUMO-SSP5. M is pre-stained protein marker and prominent bands are indicated by their size in kDa.
Secondary structure of SUMO-SSP5 was analyzed by FT-IR spectroscopy, The results showed that the conjugated protein is primarily comprised of beta sheets, as the peak was found at 1625 cm-1 (Figure 5). One of the main reasons behind the beta sheet-rich secondary structure is the presence of SSP motifs on the c-terminus. The SSP motif is comprised of three hydrophobic amino acid resides and one hydrophilic amino acid at the end. This combination of amino acids allows for non-covalent interactions, particularly, hydrogen bonding and aromatic stacking, between other copies of the motif's residues on other molecules of SUMO-SSP in a solvent exposed state. The hydrogen bonding takes place between N group of hydrophobic residues in the unfolded state of one monomer to the COO- group of residue from another monomer. The aromatic stacking mainly takes place between the side chains of phenylalanine on opposing monomers or opposing beta sheets in the same monomer in the unfolded state.
Morphological characterization of SUMO-SSPs conjugated proteins by SEM
Refolded SUMO-SSPs when observed under SEM, it was found that material was highly fibrous (Figure 5). The length of fibers was more than 100um long while the diameter was different for different SUMO-SSPs. The SUMO-SSP2 fibers were flat thick and crystalline, while SUMO-SSP1 and SUMO-SSP3 were thin and long. The morphology of material was significantly different compared to denaturing condition, which shows that upon refolding material properties change and SUMO-SSPs exhibit the fibrous structure due to monomer-monomer interaction and stacking (Figure 6-8).
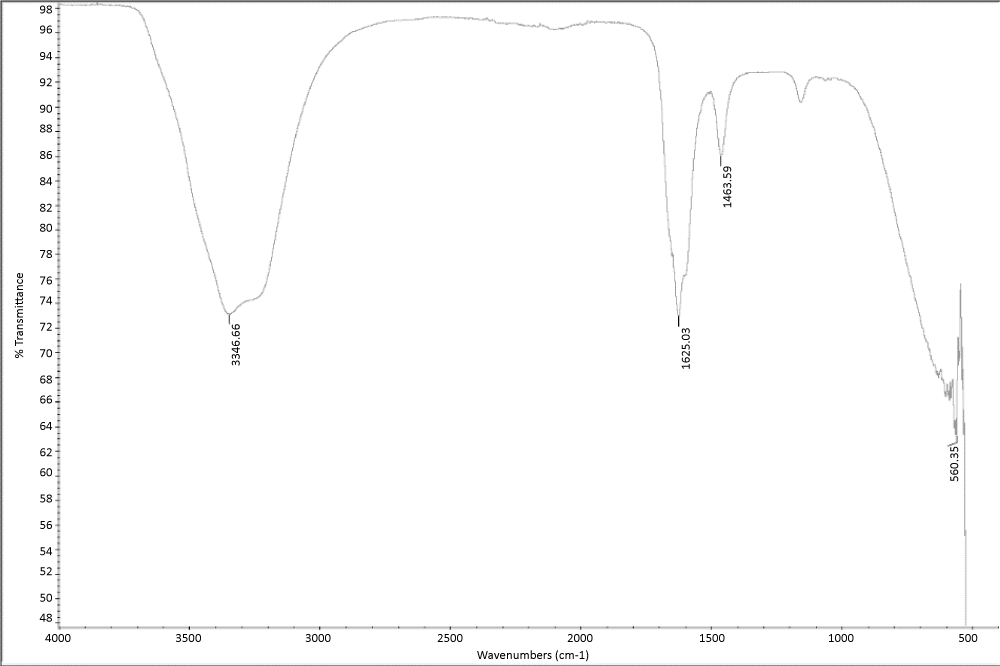
Figure 6. Secondary structure characterization of SUMO-SSP5.

Figure 7. Morphology characterization of refolded SUMO-SSPs by scanning electron microscopy. A. SUMO-SSP1 . B. SUMO-SSP2 . C. SUMO-SSP3. All refolded SUMO-SSPs have shown highly fibrous structure.
Conclusion
Many proteins aggregates due to self-association of molecules in the form of particles, precipitates and fibers and aggregation prone motifs within the protein sequence plays critical role in the association of molecules [25]. During this study, we have tried to conjugate the ultrashort self-assembling peptide motif to a highly soluble protein, SUMO, and studied its impact on the solubility of SUMO. We observed that almost all of the SUMO expression was directed to the in the inclusion bodies in the bacterial cells. This research highlights the importance of the identification of aggregation prone motifs within the protein sequences. Truncation and mutation of such motifs can improve the solubility and stability of proteins especially if the protein of interest is a biotherapeutic. However, if the target recombinant protein is toxic to the cells or to be used in the fibrous form, then such a ultrashort self-assembling peptides can be conjugated to the target protein.
Author contributions
MS and CAEH conceived the idea. MS, SSQ and HHS performed the experiment. MS and CAEH wrote the manuscript.
Funding
The research reported in this publication was supported by funding from King Abdullah University of Science and Technology (KAUST).
Conflict of Interest
The authors declare no conflicts of interest
References
- Ami D, Natalello A, Gatti-Lafranconi P, Lotti M, Doglia SM (2005) Kinetics of inclusion body formation studied in intact cells by FT-IR spectroscopy. Febs Lett 579: 3433-3436.
- Lazar GA, Marshall SA, Plecs JJ, Mayo SL, Desjarlais JR (2003) Designing proteins for therapeutic applications. Curr Opin Struct Biol 13: 513-518.
- Rinas U, Garcia-Fruitos E, Corchero JL, Vazquez E, Seras-Franzoso J, et al. (2017) Bacterial Inclusion Bodies: Discovering Their Better Half. Trends Biochem Sci 42: 726-737.
- Diener M, Kopka B, Pohl M, Jaeger KE, Krauss U (2016) Fusion of a Coiled-Coil Domain Facilitates the High-Level Production of Catalytically Active Enzyme Inclusion Bodies. Chemcatchem 8: 142-152.
- Saleem M, Moore J, Derrick JP (2012) Expression, purification, and crystallization of neisserial outer membrane proteins. Methods Mol Biol 799: 91-106.
- Villaverde A, Garcia-Fruitos E, Rinas U, Seras-Franzoso J, Kosoy A, et al. (2012) Packaging protein drugs as bacterial inclusion bodies for therapeutic applications. Microb Cell Fact 11.
- Butt TR, Edavettal SC, Hall JP, Mattern MR (2005) SUMO fusion technology for difficult-to-express proteins. Protein Expr Purif 43: 1-9.
- Peroutka Iii RJ, Orcutt SJ, Strickler JE, Butt TR (2011) SUMO fusion technology for enhanced protein expression and purification in prokaryotes and eukaryotes. Methods Mol Biol 705: 15-30.
- Roberts CJ (2014) Protein aggregation and its impact on product quality. Curr Opin Biotech 30: 211-217.
- Wang XL, Das TK, Singh SK, Kumar S (2009) Potential aggregation prone regions in biotherapeutics A survey of commercial monoclonal antibodies. Mabs 1: 254-267.
- Fink AL (1998) Protein aggregation: folding aggregates, inclusion bodies and amyloid. Fold Des 3: 9-23.
- Noble W, Hanger DP, Miller CC, Lovestone S (2013) The importance of tau phosphorylation for neurodegenerative diseases. Front Neurol 4: 83.
- Fink AL (1998) Protein aggregation: folding aggregates, inclusion bodies and amyloid. Folding & Design 3: R9-R23.
- Levine ZA, Larini L, LaPointe NE, Feinstein SC, Shea JE (2015) Regulation and aggregation of intrinsically disordered peptides. P Natl Acad Sci USA 112: 2758-2763.
- Teufel DP, Johnson CM, Lum JK, Neuweiler H (2011) Backbone-Driven Collapse in Unfolded Protein Chains. J Mol Biol 409: 250-262.
- Mao AH, Crick SL, Vitalis A, Chicoine CL, Pappu RV (2010) Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. P Natl Acad Sci USA 107: 8183-8188.
- Buck PM, Kumar S, Singh SK (2013) On the Role of Aggregation Prone Regions in Protein Evolution, Stability, and Enzymatic Catalysis: Insights from Diverse Analyses. Plos Comput Biol 9.
- Trainor K, Broom A, Meiering EM (2017) Exploring the relationships between protein sequence, structure and solubility. Curr Opin Struc Biol 42: 136-146.
- Prabakaran R, Goel D, Kumar S, Gromiha MM (2017) Aggregation prone regions in human proteome: Insights from large-scale data analyses. Proteins 85: 1099-1118.
- Chennamsetty N, Helk B, Voynov V, Kayser V, Trout BL (2009) Aggregation-Prone Motifs in Human Immunoglobulin G. J Mol Biol 391: 404-413.
- Cohen SIA, Vendruscolo M, Dobson CM, Knowles TPJ (2012) From Macroscopic Measurements to Microscopic Mechanisms of Protein Aggregation. J Mol Biol 421: 160-171.
- Carrio MM, Cubarsi R, Villaverde A (2000) Fine architecture of bacterial inclusion bodies. Febs Lett 2000 471: 7-11.
- Liovic M, Ozir M, Zavec AB, Peternel S, Komel R, et al. (2012) Inclusion bodies as potential vehicles for recombinant protein delivery into epithelial cells. Microb Cell Fact 11.
- Rodriguez-Carmona E, Cano-Garrido O, Seras-Franzoso J, Villaverde A, Garcia-Fruitos E (2010) Isolation of cell-free bacterial inclusion bodies. Microb Cell Fact 9.
- Wang X, Das TK, Singh SK, Kumar S (2009) Potential aggregation prone regions in biotherapeutics: A survey of commercial monoclonal antibodies. Mabs 1: 254-267.







