Abstract
The cMYB gene belongs to family of proto-oncogenes all of which function as strong transcriptional activators. The cMYB gene is being considered as a therapeutic target for receptor-positive breast cancer patients. Previous data show that estrogen receptor 1 (ESR1) gene regulates cMYB in MCF7 cells by regulating transcriptional elongation of the gene. These data are substantiated by knockdown studies that show that silencing ESR1 leads to downregulation of cMYB. In addition to cMYB, a substantial number of other genes are affected following silencing of ESR1. Theory is that analyses of the datasets will lead to identification of genes associated with cMYB expression, and further characterization of cMYB in breast cancers. For the current study, we analyzed two different knockdown datasets; one in which ESR1 was silenced and another in which cMYB was silenced. The goal of the study was to identify genes dysregulated because of their association with cMYB. We cross-referenced the differentially expressed genes from the ESR1 knockdown study with top candidates from a cMYB knockdown dataset, and identified LONRF2, DOK7, MSX2 and KCNMB4 genes. We suggest that these genes be considered for their contribution to our understanding of the cMYB signaling mechanisms in luminal breast cancer.
Key-words
cMYB; LONRF2, DOK7, KCNMB4; MSX2; luminal breast cancer
Introduction
The cMYB gene belongs to a family of proto-oncogenes that function as strong transcriptional activators. Thus far, of the MYB family of genes, only cMYB has validated as a bona fide oncogene in in vitro studies and is implicated in leukemia [1], colon [2], pancreatic [3], head and neck [4] and breast [5,6] cancers. Data show that cMYB is highly expressed in receptor positive luminal breast cancers, and the gene is currently being considered as a therapeutic target in breast cancers via the use of low molecular weight molecules and RNAi approaches [7]. Although their targets differ, all of the MYB family genes target genes involved in proliferation, differentiation, apoptosis and cell cycle regulation in different tissues.
Substantially more is known about cMYB compared to other MYB family genes. Studies show that cMYB can target oncogenes including MYC and KIT, suggesting that co-operative processes between the different genes may be key to driving oncogenesis [8]. Also related to its involvement in tumorigenesis, cMYB has been shown to regulate CCNB1, COX2 and MIM1, and BCL2 for a possible role in inhibition of apoptosis [6,9]. Data also show that cMYB targets cyclin b1, CCNB1, CCNA1 and CCNE1 [5,10] cell cycle genes, and GATA3, PVRL and CEBPB in breast [5]. Considered together, these data show cMYB associating with genes involved in tumor progression. The aim of this study is to further characterize cMYB in receptor positive luminal breast cancers, with the added intent of identifying genes that might eventually prove clinically useful. We retrieved and analyzed an ESR1 knock-down dataset and a cMYB knock-down dataset and identified genes common to both datasets. cMYB was substantially silenced in both datasets. From our analyses, a list of ten candidate genes were selected, one of which had been previously identified as a cMYB target. Of the ten genes, four genes validated following experimental analyses of the MCF7 model. The genes included LONRF2, DOK7, MSX2 and KCNMB4. LONRF2 is a LON peptidase N-terminal domain and the RING finger domain gene known to be involved in protein-protein and protein-DNA binding interactions associated with a number of different signaling events [11]. DOK7 (downstream of tyrosine kinase 7; docking 7) gene is essential to neuromuscular synaptogenesis and thought to activate muscle-specific receptor kinase [12]. MSX2 is a muscle homeobox gene related to regulation of bone development and regulation of signaling events between survival and apoptosis [13]. The KCNMB4 (Potassium Calcium-Activated Channel Subfamily M Regulatory Beta Subunit 4) gene is fundamental to the control of smooth muscle tone and neuronal excitability [14]. At this point, it’s unclear as to the relationship between the LONRF2, DOK7, MSX2 and KCNMB4 and cMYB. The four genes appear to be affected by conditions that silence cMYB, and further validation of their concurrent expression with cMYB should help in understanding the relationship between the signaling mechanisms.
Methods
Cell lines
The MCF7 (luminal, receptor positive) and MDA MB231 (triple negative; receptor negative) cell lines were utilized in this study. The cell lines were purchased from Atcc.org (Manassas, VA, USA) and were maintained in Dulbecco’s Modified Eagle Minimum essential media (DMEM) supplemented with 1% penicillin and 10 % serum in a 370C incubator with 5% CO2 as suggested by the supplier.
Microarray datasets obtained from Gene Expression Omnibus and data analyses
Unless noted otherwise, DNA microarray datasets used in this study were retrieved from the GEO as Affymetrix U133 plus 2 Cel Intensity files (CEL). Before uploading and utilizing the gene-chip datasets, we assessed the quality control metrics for each microarray in order to determine the quality of the hybridizations. The CEL files were uploaded into the National Cancer Institutes’ mAdb [15] web-based bioinformatics portal, normalized and the differentially expressed genes were identified following T-test and Wilcoxon group analyses and statistical filtering based on p-values of <0.05, and at least a 2-fold difference. Euclidean Hierarchical clustering (HC) was also performed using mAdb. The Maire patient sample dataset contained 101 clinical patient samples representing luminal A, luminal B and TNBC breast tissues (GSE65194) [16].
The estrogen-receptor knockdown dataset was GSM678802 [17]. The dataset was generated by siRNA mediated silencing of the estrogen receptor in MCF7 breast cancer cells. The cMYB knockdown dataset was GSE21371 [18]. cMYB was silenced following stable RNAi knockdown of endogenous cMYB. Agilent Human 4×44 K Custom Oligo microarrays (with Cy3/Cy5 channel detection) were utilized for assessment of the differentially expressed genes. We utilized the GEO2R software available via Gene Expression Omnibus to query the datasets to identify genes affected by cMYB knockdown [19]. As a default condition, the top 250 differentially expressed genes were identified based on statistical significance values <0.003. The genes identified following analyses of the Agilent custom microarray were compared to those identified using the Affymetrix microarray. The corresponding Affymetrix probe-set IDs were 204798_at for cMYB, 240633_at for DOK7, 225996_at for LONRF2 . 205555_s_at for MSX2 and 222857_s_at for the KCNMB4 gene.
RNA extraction, primer generation and PCR analyses
RNA was extracted using the TrizolTM reagent (Invitrogen; Carlsbad CA, USA) as suggested by the manufacturer. cDNA was generated for MCF7 and MDA MB231, using the iScriptTM kit (BioRad; Hercules CA, USA) as suggested by the supplier.
The primer3 program [20] was used to identify the primer sequences for each of the target genes (Table 1). The corresponding gene sequences were obtained from Affymetrix NetAffx (http://www.affymetrix.com/estore/analysis/index.affx) based on the Affymetrix probe-set ID. For each gene, the resulting amplicon sizes ranged from 200 – 300 nucleotides. The PCR primer sequences were synthesized by IDTDNA.com (Coralville Iowa, USA).
Table 1. Primer Sets for the genes of interest.
GENES |
LEFT PRIMER |
RIGHT PRIMER |
AMPLICON |
|
|
|
(base Pairs) |
DOK7 |
CMGCTCCIGTCTGAACCIC |
CC GACAGTGAAGGGACAAAG |
209 |
KCNM B4 |
ATGATGTGCITCTGCATCGC |
CO TG CCGATGAGTACAGCIT |
217 |
LONRF2 |
TGCTAGIGGAGAGIGGTGTC A |
AGGCCAGATGAGTGACCTGT |
215 |
MSX2 |
AAAGACTGCAGGAGGC AGAA |
CAGGG1TAGCAGAGCAGGAG |
317 |
cMYB |
C1TGITTGGGAGACTCTGCA |
TGCAAACACAGGATCCATGC |
227 |
GAPDH |
TCCCTGAGCTGAACGGGAAG |
GGAGGAGTGGGTGTCGCTGT |
218 |
PCRs were performed using the AmpliTaq GoldTM PCR mixture as suggested by the supplier (Life Technologies, Carlsbad CA, USA). The PCR was performed at 95 degrees for 5minutes, then 27 cycles of the following: thirty seconds at 95 degrees, thirty seconds at 58 degrees followed by 30 seconds at 78 degrees. The amplified products were analyzed on a 2% agarose gel and electrophoresed at 95volts for 30 minutes.
Results
Identification of ESR1 and cMYB knockdown in MCF7 dataset
The goal of the study was to further characterize cMYB and identify genes that appear to be reliability associated with cMYB expression. Previous data show that ESR1 can regulate cMYB gene expression in breast cancer [21]. Studies show that when ESR1 is silenced in MCF7 luminal cells, then cMYB is also affected [22]. Because we were interested in identifying cMYB-related genes, and cMYB is ‘affected’ in the ESR1 knockdown, our thought was that analyses of the ESR1 dataset would allow for detection of genes associated with cMYB.
We identified a GEO dataset in which ESR1 was silenced in MCF7 and cMYB downregulated. The mAdb web-based bioinformatics resource was utilized to identify genes differentially expressed in the control compared to the ESR1 knockdown. As comparison, we identified a MCF7 cMYB knockdown dataset, processed the genes to identify a list of differentially genes, and compared the two datasets in search for genes concordantly differentially expressed between the two knockdown studies. Ten genes were found to be differentially expressed in both datasets (Figure 1). All ten genes demonstrate a similar pattern of gene expression in both knockdown datasets. As exception, ESR1 is dramatically silenced in the ESR1 knockdown, but only marginally affected in the cMYB knockdown conditions.
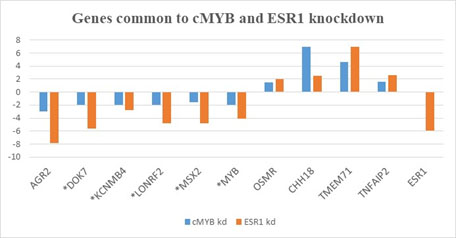
Figure 1. Genes common between the ESR1 and cMYB knockdown studies. The genes differentially expressed after cMYB knockdown were determined using GEO2R and the top 250 genes compared to genes identified as differentially expressed following ESR1 knockdown (i.e., p-value <0.05; >2.0). kd= knockdown study. Asterisk are genes that validate via PCR analyses.
Thus far, our experiments only compare the gene expression patterns. However, we have performed preliminary bioinformatics analyses to determine cMYB’s relationship to our candidate genes. Genecards and Qiagen promoter analyses [23] suggest that cMYB binds the TNFAIP2 gene promoter. Moreover, based on TF2DNA [24] analyses and Genecards, MYBL1 is suspected of binding the DOK7 gene promoter. MYBL1 and cMYB belong to the same family of proto-oncogenes, and function via reciprocal regulation [5]. It could be that cMYB regulates MYBL1, which then regulates DOK7 in luminal breast samples. The MYBL1 gene was identified in both the ESR1 and cMYB knockdown studies but was not selected (as differentially expressed) because it did not meet the statistical significance cutoff of >2.0-fold difference in the ESR1 knockdown studies. Nonetheless, the MYBL1 gene might still be functioning in the samples.
The ten genes identified above were examined via PCR using the MCF7 luminal cell line. As comparison, the gene expression levels in MDA MB231 triple negative breast cancer (TNBC) cell line were determined. Similar to cMYB, the LONRF2, DOK7, MSX2 and KCNMB4 genes consistently demonstrated a differential pattern of transcript expression. Transcript levels were highest in the MCF7 luminal sample compared to the MDA MB231 TNBC sample. As a result, these four genes are chosen as candidates that we suggest are possibly involved in signaling events consistent with cMYB (Figure 2). Cluster analyses demonstrate a gene expression pattern similar to that observed via PCR, in that transcript levels of the candidate genes and cMYB are higher in luminal compared to receptor negative TNBC patient samples (Figure 3). Approximately 76% of the luminal patient samples cluster together and 64% of the TNBC samples cluster together based on analyses of cMYB, LONRF2, DOK7, MSX2 and KCNMB4 genes. These data validate concordant gene expression between our candidate genes and cMYB.
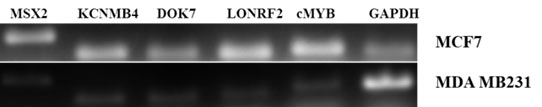
Figure 2. PCR analyses of genes common between ESR1 and cMYB knockdown datasets. MCF7 (luminal) compared to MDA MB231 (TNBC) cell lines.
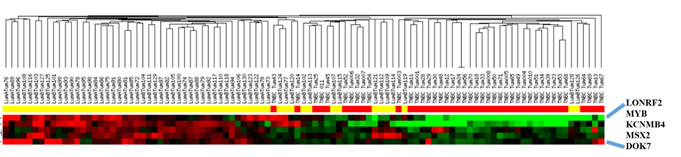
Figure 3. Hierarchical clustering of luminal compared to TNBC patient clinical samples against LONRF2, MYB, KCNMB4, MSX2 and DOK7 genes. The luminal A/B patient samples are represented by yellow and the TNBC patient samples are represented by the red clustering across the top bars.
Discussion
The cMYB gene is a strong transcriptional activator and a known oncogene. It is being considered as a therapeutic target for receptor-positive patients, hence it is the focus of numerous studies aimed at characterizing the gene. To date many of cMYB’s downstream targets have been identified. cMYB targets are involved in events and pathways key to differentiation and tumor progression [25] implicating its involvement in these signaling processes. The experiments outlined here were performed with the goal of identifying novel genes involved in cMYB signaling mechanisms. Our candidate genes are differentially expressed following ESR1/cMYB silencing and in a separate cMYB knockdown study. We reasoned that both datasets, particularly the cMYB knockdown study, should lead to identification of cMYB associated targets and interacting genes, and comparative analyses of both datasets should result in a somewhat reliable list of candidate genes.
Many of the genes identified in the ESR1 knockdown are likely driven by events associated with ESR1 targeting. So, the role of ESR1 in the events related to expression of MSX2, LONRF2, DOK7 and KCNMB4 is unclear. Data appear to suggest that ESR1 has a greater effect on cMYB, than cMYB has on ESR1 receptor. ESR1 binds to domain 1 of cMYB regulating elongation of the gene during transcription [26]. When ESR1 is knocked down there is subtantial silencing of cMYB, but comparatively when cMYB is knocked down then ESR1 is only slightly affected. Even when ESR1 is marginally affected then MSX2, LONRF2, DOK7 and KCNMB4 genes are still silenced with cMYB knockdown. It would appear that the effects on MSX2, LONRF2, DOK7 and KCNMB4 are more closely related to cMYB than ESR1.
As for the relationships of our four genes to cMYB, TF2DNA analyses of MSX2 suggest regulation of cMYB. MSX2 is a homeobox gene. As far as we are aware, gene expression has not been assessed in MCF7 cell lines, nor has levels been assessed with respect to cMYB. MSX2 has been assessed in MDA MB231 cells. MDA MB231 cells are a highly aggressive breast cancer subtype. Lanigen, et al. [27] showed that ectopic expression of MSX2 in MDA MB231 cells lead to downregulation of survivin gene and induced apoptosis. The authors did not assess cMYB expression but did substantiate low levels of MSX2 in MDA MB231 which also express low endogenous levels of cMYB.
The DOK7 gene functions in synaptogenesis, mostly described in processes associated with neuronal cell types with limited studies relating the gene to cancers. Studies of DOK7 show the gene is regulated by methylation with reduced levels in lung [28] and breast [29], with reduced expression associated with poor prognosis. The gene does not appear to be directly related to cMYB, however, TF2DNA analyses suggest DOK7 can be regulated by MYBL1. MYBL1 is a member of the cMYB family and is expressed in some luminal breast including MCF7. It could be that DOK7 association in MCF7 is via regulation by MYBL1. DOK7 is not detected in MDA MB231, however MYBL1 is expressed in the cell lines [30], suggesting a possible different method of regulation in MDA MB231 cells.
The LONRF2 gene is identified as being one of a list of coding genes differentially expressed in both colon and breast cancers [31], with only one citation related to breast and overall limited analyses. As far as we are aware the KCNMB4 gene has not been described in either cancers or breast.
Conclusion
This study is intended to be a preliminary analysis aimed at identifying genes related to the cMYB gene. We have identified four genes that appear to be related to responses either directly or indirectly associated with cMYB expression in MCF7 and luminal patient samples. In addition to identifying the candidate genes, our laboratory is interested in understanding the role of these genes in breast cancers.
Acknowledgments
This study was supported in equal parts by the Summer Undergraduate Research Program (SURP) of the College of Science Technology and Engineering (TL and GT) and the Department of Biology at Texas Southern University (NA). AP would also like to thank the mAdb Bioinformatics Center at the National Cancer Institutes for their continued support, allowing her to use their data analyses resources.
Larkins T and Tavera G equally contributed to the work.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Jin S, Zhao H, Yi Y, Nakata Y, Kalota A, et al. (2010) c-Myb binds MLL through menin in human leukemia cells and is an important driver of MLL-associated leukemogenesis. J Clin Invest 120: 593-606. [Crossref]
- Wilkins HR, Doucet K, Duke V, Morra A, Johnson N (2010) Estrogen prevents sustained COLO-205 human colon cancer cell growth by inducing apoptosis, decreasing c-myb protein, and decreasing transcription of the anti-apoptotic protein bcl-2. Tumour Biol 31: 16-22. [Crossref]
- Azim S, Zubair H, Srivastava SK, Bhardwaj A, Zubair A, et al. (2016) Deep sequencing and in silico analyses identify MYB-regulated gene networks and signaling pathways in pancreatic cancer. Sci Rep 6: 28446. [Crossref]
2021 Copyright OAT. All rights reserv
- Rettig EM, Tan M, Ling S, Yonescu R, Bishop JA, et al. (2015) MYB rearrangement and clinicopathologic characteristics in head and neck adenoid cystic carcinoma. Laryngoscope 125: E292-E299. [Crossref]
- Rushton JJ, Davis LM, Lei W, Mo X, Leutz A, et al. (2003) Distinct changes in gene expression induced by A-Myb, B-Myb and c-Myb proteins. Oncogene 22: 308-313. [Crossref]
- Ramsay RG, Gonda TJ (2008) MYB function in normal and cancer cells. Nat Rev Cancer 8: 523-534. [Crossref]
- Zhou Y, Ness SA (2011) Myb proteins: angels and demons in normal and transformed cells. Front Biosci (Landmark Ed) 16: 1109-11031. [Crossref]
- Liu X, Xu Y, Han L, Yi Y (2018) Reassessing the Potential of Myb-targeted Anti-cancer Therapy. J Cancer 9: 1259-1266. [Crossref]
- Mitra P, Yang R, Sutton J, Ramsay RG, Gonda TJ (2016) CDK9 inhibitors selectively target estrogen receptor-positive breast cancer cells through combined inhibition of MYB and MCL-1 expression. Oncotarget 7: 9069-9083. [Crossref]
- Werwein E, Schmedt T, Hoffmann H, Usadel C, Obermann N, et al. (2012) B-Myb promotes S-phase independently of its sequence-specific DNA binding activity and interacts with polymerase delta-interacting protein 1 (Pdip1). Cell Cycle 11: 4047-4058. [Crossref]
- Hua Y, Ma X, Liu X, Yuan X, Qin H, et al. (2017) Abnormal expression of mRNA, microRNA alteration and aberrant DNA methylation patterns in rectal adenocarcinoma. PLoS One 12: e0174461. [Crossref]
- Okada K, Inoue A, Okada M, Murata Y, Kakuta S, et al. (2006) The muscle protein Dok-7 is essential for neuromuscular synaptogenesis. Science 312: 1802-1805. [Crossref]
- Wu Q, Zhang L, Su P, Lei X, Liu X, et al. (2015) MSX2 mediates entry of human pluripotent stem cells into mesendoderm by simultaneously suppressing SOX2 and activating NODAL signaling. Cell Res 25: 1314-1332. [Crossref]
- Behrens R, Nolting A, Reimann F, Schwarz M, Waldschütz R, et al. (2000.) hKCNMB3 and hKCNMB4, cloning and characterization of two members of the large-conductance calcium-activated potassium channel beta subunit family. FEBS Lett 474: 99-106. [Crossref]
- Greene JM, Asaki E, Bian X, Bock C, Castillo S, et al. (2003) The NCI/CIT microArray database (mAdb) system - bioinformatics for the management and analysis of Affymetrix and spotted gene expression microarrays. AMIA Annu Symp Proc 1066. [Crossref]
- Maire V, Baldeyron C, Richardson M, Tesson B, Vincent-Salomon A, et al. (2013) TTK/hMPS1 is an attractive therapeutic target for triple-negative breast cancer. PLoS One 8: e63712. [Crossref]
- Al Saleh S, Al Mulla F, Luqmani YA (2011) Estrogen receptor silencing induces epithelial to mesenchymal transition in human breast cancer cells. PLoS One 6: e20610. [Crossref]
- Thorner AR, Parker JS, Hoadley KA, Perou CM (2010) Potential tumor suppressor role for the c-Myb oncogene in luminal breast cancer. PLoS One 5: e13073. [Crossref]
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, et al. (2013) NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res 41: D991-D995. [Crossref]
- Koressaar T, Remm M (2007) Enhancements and modifications of primer design program Primer3. Bioinformatics 23: 1289-1291. [Crossref]
- Guerin M, Sheng ZM, Andrieu N, Riou G (1990) Strong association between c-myb and oestrogen-receptor expression in human breast cancer. Oncogene 5: 131-135. [Crossref]
- Lei W, Rushton JJ, Davis LM, Liu F, Ness SA (2004) Positive and negative determinants of target gene specificity in myb transcription factors. J Biol Chem 279: 29519-29527. [Crossref]
- Safran M, Chalifa-Caspi V, Shmueli O, Olender T, Lapidot M, et al. (2003) Human Gene-Centric Databases at the Weizmann Institute of Science: GeneCards, UDB, CroW 21 and HORDE. Nucleic Acids Res 31: 142-146. [Crossref]
- Pujato M, Kieken F, Skiles AA, Tapinos N, Fiser A, et al. (2014) Prediction of DNA binding motifs from 3D models of transcription factors; identifying TLX3 regulated genes. Nucleic Acids Res 42: 13500-13512. [Crossref]
- Ness SA (2003) Myb protein specificity: evidence of a context-specific transcription factor code. Blood Cells Mol Dis 31: 192-200. [Crossref]
- Drabsch Y, Hugo H, Zhang R, Dowhan DH, Miao YR, et al. (2007) Mechanism of and requirement for estrogen-regulated MYB expression in estrogen-receptor-positive breast cancer cells. Proc Natl Acad Sci U S A 104: 13762-13767. [Crossref]
- Lanigan F, Gremel G, Hughes R, Brennan DJ, Martin F, et al. (2010) Homeobox transcription factor muscle segment homeobox 2 (Msx2) correlates with good prognosis in breast cancer patients and induces apoptosis in vitro. Breast Cancer Res 12: R59. [Crossref]
- Chen G, Yu H, Satherley L, Zabkiewicz C, Resaul J, et al. (2017) The downstream of tyrosine kinase 7 is reduced in lung cancer and is associated with poor survival of patients with lung cancer. Oncol Rep 37: 2695-2701. [Crossref]
- Heyn H, Carmona FJ, Gomez A, Ferreira HJ, Bell JT, et al. (2013) DNA methylation profiling in breast cancer discordant identical twins identifies DOK7 as novel epigenetic biomarker. Carcinogenesis 34: 102-108. [Crossref]
- Player A, Abraham N, Burrell K, Bengone IO, Harris A, et al. (2017) Identification of candidate genes associated with triple negative breast cancer. Genes Cancer 2017. 8: 659-672. [Crossref]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, et al. (2006) The consensus coding sequences of human breast and colorectal cancers. Science 314: 268-274. [Crossref]



