Three-dimensional (3D) cell culture models became an important technique in oncology research. These novel 3D techniques permit the investigation of the tumor microenvironment, promising clinical assays to predict patient response and enhanced drug selection for clinical trials. Although early diagnosis and development of new treatment strategies led to improved prognosis, the burden for the patient still remains high, without guaranteed achievement.
These biological complex, but necessary models have already been shown significant advancements and need to be narrowly enrolled in cancer drug discovery in the future. Here, the human tumor slice culture is shortly presented with shortcomings, advantages and points to consider, when handling this simple but complex 3D tumor model.
For many decades, preclinical cancer research has relied on 2D cell culture models to identify novel drug candidates for cancer treatment, followed by testing the activity of the compounds in murine tumor xenograft models. Unfortunately, promising results from cell culture and animal experiments often do not correlate with the clinical efficacy of the respective compounds in humans. On the other hand, prediction of the efficacy of established treatment approaches represents an important field of research, since different patients do not respond to the same compounds in the same way. A number of predictive biomarkers have been developed during recent years, which are now routinely used in the clinical setting, mostly in the area of targeted therapies. Only few biomarkers which are in clinical use ensure a highly accurate prediction of treatment response. Still, assays to predict an individual patient´s response towards a distinct treatment approach (i.e. chemotherapy, targeted therapy, immunotherapy etc.) are urgently needed for optimal cancer treatment in the era of "personalized medicine".
Human tissue models
Standard research models like tumor xenografts and monolayer cell cultures have evident shortcomings in the light of recent cancer research. Species differences and cellular alterations in cell lines are only two examples of profound limitations. In addition, oncological research is focusing more and more on extracellular matrix, cross-talk between cancer cells and the tumor micro milieu, including immunological conditions. Several system models, to investigate these new fields, evolved. Two particularly promising methods are organoid spheroids and tissue slice cultures. Spheroids, grown from tumor stem cells or tumor pieces are organized structures that comprise various differentiated cell types and can be expanded without apparent limitation. Slice cultures are thin cuts of the original tumor tissue, preserving the original morphology and cellular composition of tumors for several days up to weeks in culture, depending on the tumor entity (Figure 1a-d). While spheroid models are widely described and recently reviewed for intestinal cancers [1] and others [2], we focus here on a brief overview of the slice culture model and describe future possibilities in this field.
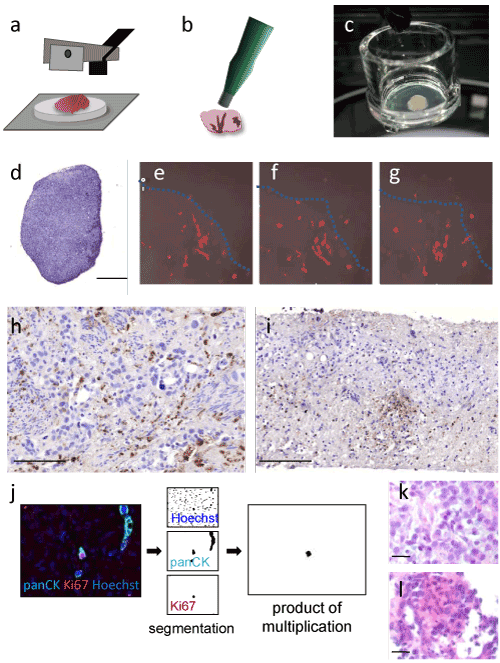
Figure 1: Human tumor slice cultures. (a) Defined (350µm) slices of the tumor were obtained by a tissue chopper (McIlwain TC752; Campden Instruments, USA). (b) After tissue slice were separated, the diameter of each tissue slice was standardized by using a manual 3mm coring tool (kai Europe, Solingen, Germany). (c) A coring tool shaped tumor slice after three days in culture on an insert membrane. Only the lower side of the tissue is in contact with the culture media, the upper side stays exposed to air. (d) HE staining of a gastric cancer slice, cultivated for four days. (e-g) Co-culture of gastric cancer tissue (upper side) and adjacent gastric tissue treated with an iron scavenger (lower side). Adjacent tissue was cultured for 24h with Isolectin B4 (647 labelled) and was then transferred to the tumor slice culture without macrophage labelling. Tissue was live imaged by a confocal microscope with a stable incubation chamber and a 20x objective. (e) One, (f) two and (g) three hours after initial co-culture. (h) CD68 (macrophage) DAB staining of a horizontal section of a gastric cancer slice culture after four days in culture. (i) CD3 (lymphocyte) DAB staining of a vertical section of a gastric cancer slice culture after six days in culture. (j) ImageJ based analysis of Ki67 positive gastrointestinal tumor cells (pan-cytokeratin, AE1-3), counterstained with Hoechst 33342 to evaluate drug impact on tumor proliferation. (k) HE staining of gastric cancer biopsy tissue directly fixed after endoscopy and (l) after four days in culture under control conditions. Bars: (d) 500µm; (h,i) 100µm; (k,l) 20µm
Human tumor slice cultures – an enhanced system model
Tumor slice cultures can be useful for studying complex cellular processes and tumor-stroma interaction ex-vivo. In 1997 it was already shown that antibodies against a cell-surface receptor altered the growth pattern of cancerous cells kept in 3D cultures but not in monolayer cultures [3]. Since then, numerous changes were discovered and novel systems became established (reviewed in [4,5]). It became clear that tumor tissue consists of various cancer cell clones, influencing each other. These compilation needs to be maintained similar to the physiological condition, in order to compare tissue response in culture with the tumor response of the organism [6,7]. Intriguing papers were published that report about investigation and refinement of various technological issues. Vaira et al. and Unger et al. could describe a good clonal maintenance of cultured lung, colon and prostate tumor slice tissue in comparison to tissue collected directly after surgery [8,9]. Colon cancer tissue of the same patients was used to compare slice cultures ex vivo with primary monolayer cultures by microarray analysis. Colon tumor slice cultures showed little alterations whereas primary cultures were not comparable to the uncultured tumor tissue. However, an important point to consider in this comparison is the different culture media used in the compared methods [9]. Moreover, promising novel therapeutics and mechanisms of resistance can be tested on human tumor slice cultures, guiding and potentially even preventing animal experiments. Animal research in cancer has profound impact on the quality of life of the animals and they may lead to severe suffering and death of the animal. Besides mutational mouse models, xenograft models are scientifically questionable in the context of tumor-stroma interaction. These mouse models are the golden standard in oncology research, yet they should be used with care as not only a missing immune-system but also species differences need to be considered. That species differences can have a severe impact was seen in the so-called ‘London tragedy’. Fatal cytokine storms were developed by all human volunteers taking part in a clinical phase I trial. The investigational compound, a monoclonal antibody directed against Cluster-of-differentiation (CD) 28 did not indicate any of these effects in non-human chimpanzees or mouse models. CD28 is one of the proteins expressed on human T cells that provide co-stimulatory signals required for T cell activation and survival. In contrast to the animal models, the reason for the observed severe clinical effect was reproducible in human lymphatic tissue slice cultures from the palentine tonsil. The obtained slices were treated with the monoclonal antibody against CD28 and the culture media was monitored for cytokine release for 72h (unpublished). Later, the antibody was identified to be a super-agonist in humans and a clinical trial using very small doses indicates now first signs of clinical efficacy, when applied in patients with rheumatoid arthritis [10].
In summary, the human tumor slice culture model is an enhanced model to study tumor heterogeneity ex-vivo and has the potential to reduce the translational step towards clinical studies.
Tumor tissue slice culture –technical considerations
Until today, slice cultures have mainly been used and are mainly published to test susceptibility to anti-cancer drugs [11-16]. In Glioblastoma slice cultures, the clinical O(6)-methylguanine-DNA methyltransferase (MGMT) promoter methylation did not reflect tumor slice culture response to clinical standard treatment. This, however, is also seen in clinical observations [11]. Further studies investigated tumor susceptibility in tumor slice cultures, however, the promising results are not yet clinically approved [8,12,13,15]. Thus, conserving tissue architecture and cellular diversity ex-vivo tumor slice culture promises to be a representative tumor model.
The group from the innovative Medicines Initiative Consortium ‘PREDECT’ analyzed in depth different culture conditions of various xenografted tumors, supporting the cultivation on a liquid-air interface above floating cultures [17]. The different aspects of these approaches should be carefully considered and future research will demonstrate advantages and disadvantages of each set-up. One major advantage of tumor slice cultures on membranes is the possibility to image cellular activities live with a confocal microscope (Figure 1e-g). Studies using membranes to cultivate tumor slice cultures on a liquid-air interface could keep glioblastoma tissue slice cultures 16 days in culture [11]. Gastric cancer, colon cancer, head and neck tumor and non-small lung cancer slice cultures are well maintained up to six or seven days ex vivo [12,13]. Considering the survival of all cellular layers, 350-400µm seem to be the most suitable thickness for tumor slice cultures while 300-350µm are primarily used for tumor tissue and conventional organotypic hippocampal slice cultures. Tissue cultures below 200µm are difficult to maintain, as the tissue architecture diverges. Above 400µm thick tissue cultures cannot provide the nutritional and oxygen need throughout the complete tissue.
Even if the tissue is damaged and inflammatory processes are provoked at the surface of the tissue slice, cells within cultured tissue need simply to adapt to metabolic changes, depending on the culture media. These culture media are however detrimental as the media remain one of the main factors that need to be controlled closely to obtain reliable results. We tested some different basic media but did not observe obvious changes regarding the cultivation of tumor slice cultures. But we assume that supplementary growth factors and media composition will have some impact on tissue integrity and survival of certain cell populations. Although tissue slices were cultivated floating and not at a liquid-air interface, Naipal et al. could show in breast cancer tissue slices effects of different medium conditions, demonstrating enhanced survival of the tissue in culture media with only two percent fetel calf serum and defined supplements [15]. Majumder et al. set up an elegant tissue-matrix system to cultivate tissue specimens; they showed that the supplementation of 2% autologous serum to culture media enhanced ATP utilization and proliferation of the tissue. Moreover they observed a similar activation profile of some main receptor tyrosine kinases [18]. Individual serum, however also may carry concentrations of various therapeutic substances depending on individual medication. Therefore, it is also a matter of experimental question whether the media should be fully defined or supplemented with animal or even individual human serum. Defined culture media might even prolong the culture period of some tumor entities and enhance the survival of cellular populations within the tissue. This issue was already pointed out by Unger et al. in 2014 [19]. However, nutritional uptake, oxygen supply and the influence of culture media on tumor slice cultures are still poorly explored.
Very few immunohistochemsitry (IHC) stainings demonstrate intact vessels in formalin-fixed, paraffin-embedded (FFPE) sections of tumor slice cultures [8]. However, Davis et al. observed in slice cultures of non-small lung cancer xenografts hypoxic areas. These areas are observed in the tissue culture at the membrane-facing side. While this obvious limitation of the model urges for detailed investigation, the interesting point of tumor cells surviving hypoxic conditions might become a novel aspect of the tumor slice culture model. Still, the vasculature in tumor slice cultures needs to be closely investigated together with possible perfusion systems, not yet established in common tumor slice cultures. These facts are detrimental, regarding drug delivery and uptake as most studies dissolve the drug of investigation in culture media.
In sum we can say that even if diverse approaches are used in different studies, important considerations emerge and need to be addressed in future studies. More standardized research is needed to strengthen reproducible results and highlight the hallmarks of human tumor slice culture.
Tumor tissue slice cultures- analysis
Human tissue is scarce and precious, therefore we carefully need to select experiments the tissue is most valuable for. The effort to develop a potent model for personalized medicine should be simultaneously designed to improve methodological questions and to study principal mechanisms of tumor heterogeneity and drug resistance. The thus far established applications offer possibilities that might be valuable for future drug development; even high-throughput screening will not be possible with this methodological approach. Although the tissue is limited common analysis systems are applicable to determine susceptibility. Histological analysis is time-consuming, but still of remarkable importance within the system of tumor slice cultures. Most investigators use proliferating markers, like Ki67 or 5-ethynyl-2'-deoxyuridine (proliferation kit) in combination with histological tumor markers or morphological analysis to study tumor susceptibility. Common pharmacological approaches however are not suitable, as they cannot control for cellular characteristics. Selection of the FFPE sections is crucial to obtain reliable results. FFPE sections were carefully selected for establishing a free, pixel-based and semi-automated read out (Figure 1d). This image J based method and similar commercially available systems enable to obtain reliable results within one week after surgery or biopsy (Figure 1j). Considering a personalized medicine approach, a fast and observer-independent read-out is necessary, as results are obtained in order to influence the decision making process for real-life therapeutic interventions. To further develop diagnostic tools for tumor slice cultures, histopathological expertise and correlation is definitely needed. Molecular analysis of tumors that are resistant to therapeutic interventions might indicate novel targets. Comparing these data with the untreated, initial tumor sample, might then indicate predictive markers. But also the investigation of culture media can be examined to monitor metabolic changes and markers that correspond with observed histological responses ex-vivo and, more importantly, in-vivo [20].
Tumor slice cultures- future perspectives
The treatment of cancer patients is based on chemotherapy, surgery, radiation, monoclonal antibodies and since recently immune checkpoint regulators. All drug therapies and combinations can be examined in slice cultures of material obtained during surgery and prospective correlation of tissue slice culture data with clinical outcomes is soon expected [11,12]. However, surgery, where the necessary tissue for personalized treatment testing is collected, is not the first treatment approach for all cancer types. Therefore, biopsy samples need to be examined whether they are suitable for tissue cultures and subsequently can indicate treatment responses. For example, gastrointestinal cancers are heterogeneous and aggressive tumors, that often respond poorly to many standard chemotherapies or inevitably relapse after initial response [21-23]. All these facts point to tumor heterogeneity and evolution during treatment. As biopsies yield limited tissue samples, it needs to be considered that ex-vivo drug testing in one selected tissue probe may not represent the whole molecular make-up of the tumor. First experiments in our laboratory, however, show promising effects (Figure 1k,l).
Newly applied immune stimulating agents (“immune checkpoint inhibitors and beyond”) might provoke cytokine release syndromes in cancer patients, but still no system model seems to be able to predict the effectiveness or the potentially dangerous side-effects of this group of drugs [24]. Slice cultures of tumor tissue bear potential in this regard, as tumor tissue can be cultured in its integrity and even can be co-cultured with lymphoid tissue or adjacent healthy tissue (Figure 1e-g). Simple high density cultures of lymphocytes from peripheral blood mononuclear cells are identified to be a suitable model for human lymphocyte cultures, mimicking physiological markers [25]. These cultures can be easily combined with tumor slice cultures, obtaining an adequate model to study immunoregulators in a human setting. Although the tumor slice cultures are immune-competent by themselves (Figure 1h,i), not only co-culture of tumor and immune cells are of interest [12]. Co-culture of tumor tissue with adjacent healthy tissue enables various approaches to identify and verify upcoming players to combat cancer. For example the central role of macrophages, shaping the extracellular matrix can now be investigated in a more relevant system model, seen in Figure 1e-g. However, tumor slice cultures need to be carefully evaluated for immune competence, since the activation state of the immune system has important influence on drug susceptibility. The identification and differentiation of beneficial and harmful immunoreactions will be essential. Further approaches may consider clinically applicable transduction and transfection models, like adeno-associated viral vectors or nanoparticles. Moreover, novel promising viral applications could be initially tested ex-vivo [26-28]. Especially in cancer, where viral infection can initiate carcinogenesis, human 3-D cultures of healthy tissue are highly of advantage [29]. Novel model systems, like long term mini gut cultures, derived from human biopsy samples co-cultured with tumor slices cultures are also highly interesting to investigate tumor growth as it has been observed that healthy gastric tissue alone cannot be cultured ex vivo over a longer period [30,31].
Future developments, like the ambitious ‘tumor on a chip’ model allows for cultivation of several system models, tissues, organoids, cellular cultures with a functional perfusion system on a scale of a conventional slide. This approach already meets the need of stabilized nutritional supply and enhanced possibilities of co-cultures [32,33]. This technique is fascinating as the drug response of all organs of the organism can be considered and examined. However, only few laboratories are able to afford the still exquisite system and the small amount of tissue may not meet the aspect of heterogeneity in cancer. Still, advances in membrane systems and incubation possibilities promise further improvements for tissue integrity and may enable stable and long-time tissue cultures [17,34].
In summary, major advantages of human tissue slice cultures are the maintenance of tissue integrity as well as the direct possibility to observe individual tumor susceptibility and resistance mechanisms. Major limitations are the lack of a functional vasculature, limited culture periods and that the method is not suitable for high throughput approaches. To the best of our knowledge, tumor slice cultures are not yet used in drug development and more research is needed to develop standardized, fast and robust readout technologies. Nevertheless, human tumor slice cultures are a promising approach for clinical application and represent a human model that narrows the gap between monolayer cell experiments, animal studies and complex clinical studies in many fields of tumor research.
This work was funded by the German Ministry of Education and Research (BMBF) Grant (FKZ 031A579)
- Short SP, Costacurta PW, Williams CS (2017)Using 3D Organoid Cultures to Model Intestinal Physiology and Colorectal Cancer. Curr Colorectal Cancer Rep 13: 183-191.
- Weiswald LB, Bellet D, Dangles-Marie V (2015) Spherical Cancer Models in Tumor Biology. Neoplasia 17: 1-15. [Crossref]
- Weaver VM1, Petersen OW, Wang F, Larabell CA, Briand P, et al. (1997) Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol 137: 231-245. [Crossref]
- Asghar W, El Assal R, Shafiee H, Pitteri S, Paulmurugan R, et al. (2015) Engineering cancer microenvironments for in vitro 3-D tumor models. Mater Today 18: 539-553. [Crossref]
- Fong EL, Harrington DA, Farach-Carson MC, Yu H (2016) Heralding a new paradigm in 3D tumor modeling. Biomaterials 108: 197-213. [Crossref]
- Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, et al. (2015) Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med 21: 795-801. [Crossref]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, et al. (2013) Cancer genome landscapes. Science 339: 1546-1558. [Crossref]
- Vaira V, Fedele G, Pyne S, Fasoli E, Zadra G, et al. (2010) Preclinical model of organotypic culture for pharmacodynamic profiling of human tumors. Proc Natl Acad Sci U S A 107: 8352-8356. [Crossref]
2021 Copyright OAT. All rights reserv
- Unger FT, Bentz S, Krüger J, Rosenbrock C, Schaller J, et al. (2015) Precision Cut Cancer Tissue Slices in Anti-Cancer Drug Testing. J Mol Pathophysiol 4: 108.
- Tyrsin D, Chuvpilo S, Matskevich A, Nemenov D, Römer PS, et al. (2016) From TGN1412 to TAB08: the return of CD28 superagonist therapy to clinical development for the treatment of rheumatoid arthritis. Clin Exp Rheumatol 34: 45-48. [Crossref]
- Merz F, Gaunitz F, Dehghani F, Renner C, Meixensberger J, et al. (2013) Organotypic slice cultures of human glioblastoma reveal different susceptibilities to treatments. Neuro-Oncol 15: 670-681. [Crossref]
- Gerlach MM, Merz F, Wichmann G, Kubick C, Wittekind C, et al. (2014) Slice cultures from head and neck squamous cell carcinoma: a novel test system for drug susceptibility and mechanisms of resistance. Br J Cancer 110: 479-488. [Crossref]
- Koerfer J, Kallendrusch S, Merz F, Wittekind C, Kubick C, et al. (2016) Organotypic slice cultures of human gastric and esophagogastric junction cancer. Cancer Med 5: 1444-1453. [Crossref]
- Faversani A, Vaira V, Moro GP, Tosi D, Lopergolo A, et al. (2014) Survivin family proteins as novel molecular determinants of doxorubicin resistance in organotypic human breast tumors. Breast Cancer Res 16: R55. [Crossref]
- Naipal KA, Verkaik NS, Sánchez H, van Deurzen CH, den Bakker MA, et al. (2016) Tumor slice culture system to assess drug response of primary breast cancer. BMC Cancer 16: 78. [Crossref]
- Holliday DL, Moss MA, Pollock S, Lane S, Shaaban AM, et al. (2013) The practicalities of using tissue slices as preclinical organotypic breast cancer models. J. Clin. Pathol 66: 253-255. [Crossref]
- Davies EJ, Dong M, Gutekunst M, Närhi K, van Zoggel HJ, et al. (2015) Capturing complex tumour biology in vitro: histological and molecular characterisation of precision cut slices. Sci Rep 5: 17187. [Crossref]
- Majumder B, Baraneedharan U, Thiyagarajan S, Radhakrishnan P, Narasimhan H, et al. (2015) Predicting clinical response to anticancer drugs using an ex vivo platform that captures tumour heterogeneity. Nat Commun 6: 6169. [Crossref]
- Unger C, Kramer N, Walzl A, Scherzer M, Hengstschläger M, et al. (2014) Modeling human carcinomas: Physiologically relevant 3D models to improve anti-cancer drug development. Adv Drug Deliv Rev: 50-67. [Crossref]
- Siravegna G, Marsoni S, Siena S, Bardelli A (2017) Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 14: 531-548. [Crossref]
- Lordick F, Lorenzen S, Yamada Y, Ilson D (2014) Optimal chemotherapy for advanced gastric cancer: is there a global consensus? Gastric Cancer Assoc 17: 213–225. [Crossref]
- Van Cutsem E1, Köhne CH, Hitre E, Zaluski J, Chang Chien CR, et al. (2009) Cetuximab and Chemotherapy as Initial Treatment for Metastatic Colorectal Cancer. N Engl J Med 360: 1408-1417. [Crossref]
- Pietrantonio F, Vernieri C, Siravegna G, Mennitto A, Berenato R, et al. (2017) Heterogeneity of Acquired Resistance to Anti-EGFR Monoclonal Antibodies in Patients with Metastatic Colorectal Cancer. Clin Cancer Res 23: 2414-2422. [Crossref]
- Kroschinsky F, Stölzel F, von Bonin S, Beutel G, Kochanek M, et al. (2017) New drugs, new toxicities: severe side effects of modern targeted and immunotherapy of cancer and their management. Crit Care 21: 89. [Crossref]
- Wegner J, Hackenberg S, Scholz CJ, Chuvpilo S, Tyrsin D, et al. (2015) High-density preculture of PBMCs restores defective sensitivity of circulating CD8 T cells to virus- and tumor-derived antigens. Blood 126: 185-194. [Crossref]
- Kallendrusch S, Schopow N, Stadler SC, Büning H, Hacker UT (2016) Adeno-Associated Viral Vectors Transduce Mature Human Adipocytes in Three-Dimensional Slice Cultures. Hum Gene Ther Methods 27: 171-173. [Crossref]
- Merz L, Höbel S, Kallendrusch S, Ewe A, Bechmann I, et al. (2017) Tumor tissue slice cultures as a platform for analyzing tissue-penetration and biological activities of nanoparticles. Eur J Pharm Biopharm 112: 45-50. [Crossref]
- Marino N, Illingworth S, Kodialbail P, Patel A, Calderon H, et al. (2017) Development of a versatile oncolytic virus platform for local intra-tumoural expression of therapeutic transgenes. PLoS One 12: e0177810. [Crossref]
- Burkitt MD, Duckworth CA, Williams JM, Pritchard DM (2017) Helicobacter pylori -induced gastric pathology: insights from in vivo and ex vivo models. Dis Model Mech 10: 89-104. [Crossref]
- Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, et al. (2011) Long-term Expansion of Epithelial Organoids From Human Colon, Adenoma, Adenocarcinoma, and Barrett’s Epithelium. Gastroenterology 141: 1762-1772. [Crossref]
- VanDussen KL, Marinshaw JM, Shaikh N, Miyoshi H, Moon C, et al. (2015) Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut 64: 911-920. [Crossref]
- Albanese A, Lam AK, Sykes EA, Rocheleau JV, Chan WC (2013) Tumour-on-a-chip provides an optical window into nanoparticle tissue transport. Nat Commun 4: 2718. [Crossref]
- Ahn J, Sei Y, Jeon N, Kim Y (2017) Tumor Microenvironment on a Chip: The Progress and Future Perspective. Bioengineering 4: 64.
- Kallendrusch S, Merz F, Bechmann I, Mayr SG, Zink M (2017) Long-Term Tissue Culture of Adult Brain and Spleen Slices on Nanostructured Scaffolds. Adv Healthc Mater 6: 1601336. [Crossref]

