The widespread resistance to carbapenem via Enterobacteriaceae is a major public health concern. To investigate the reason for the change of Escherichia coli from carbapenem-susceptible to carbapenem-resistant status after antibiotic therapy, whole-genome sequencing was applied for the analysis of genetic difference between the two isolates. The genome of the two isolates of Escherichia coli with distinct resistant phenotypes recovered from one patient was sequenced and compared. Single-nucleotide polymorphisms and insertions/deletions were analyzed using software MUMmer (http://mummer.sourceforge.net/). The homology of chromosomes, plasmid 1, and plasmid 2 between the two isolates was very high (>99.5%). However, the coverage ratio of plasmid 3 was only 7.13%. This plasmid harbored the IMP-4 carbapenemase-coding gene. The upstream of blaIMP-4 was the plasmid mobilization relaxosome protein Mobe gene and the downstream were class 1 integrom integrase IntI1 and IS6 family transposase IS15DIV, which may be related to carbapenemase gene capture. Single-nucleotide mutation, insertion, and deletion could not result in the development of resistance to carbapenem, whereas acquisition of the IMP-4-encoding gene may be the reason for changes in resistance to carbapenem.
carbapenem-resistant Enterobacteriaceae, whole-genome sequencing analysis, plasmid-encoded carbapenemase
The abundance of multidrug-resistant Gram-negative bacteria has increased alarmingly in the last two decades worldwide. In particular, the emergence of carbapenem-resistant Enterobacteriaceae (CRE) has become a new challenge in the treatment of infectious diseases [1,2]. In China, the isolation ratio of CRE has increased five-fold from 2005 to 2014 [3]. The mechanism of resistance to carbapenem in Enterobacteriaceae is mainly mediated by the production of carbapenemases, which are capable of hydrolyzing the carbapenems, and production of an extended-spectrum β-lactamase (ESBL) and/or AmpC cephalosporinase (AmpC) in conjunction with membrane impermeability or active drug efflux pumps [4-6]. CRE is difficult to control in the clinical setting because of the multiple acquisition pathways. These include the endogenous pathway through antibiotic selective pressure on intestinal microbiota and exogenous pathway through horizontal transmission or through a combination of these factors [5]. Responding to the threat of antimicrobial resistance (AMR), which includes surveillance and stewardship, has been designated as a strategic priority worldwide. For improved monitoring, detection, and screening of AMR, it is important to determine the AMR genes that are prevalent, genes that are moving around, and those that pose the greatest threat. In recent years, whole-genome sequencing (WGS) has been gradually used for the genotype-based diagnosis of AMR [7-9]. WGS has great potential for epidemiological tracking and understanding the development of resistance via experimental evolution. DNA analysis also offers the opportunity to construct databases that record genes of interest, the mobile elements that move these genes, and the cells or species that acquire such genes [10].
In this study, two isolates of Escherichia coli (E. coli) were collected from one patient prior to and after antimicrobial therapy; one isolate was susceptible to carbapenem, whereas the other was resistant. We performed WGS to identify the genetic heterogeneity within the isolates that result in change of resistance to carbapenem.
Patient and bacterial isolates
One isolate of E. coli E41-2, which was susceptible to carbapenem, was recovered from sputum. After 10 days of antimicrobial therapy, a carbapenem-resistant E. coli E41-1 was also isolated. The isolates were identified by Vitek2-Compact. The minimum inhibitory concentration of the two isolates to antibiotics is listed in Table 1. The crude enzyme extracts of the two isolates were gathered using the repetitive freeze-thawing method. Imipenem hydration activity was determined through the modified Hodge test.
Table 1. Antimicrobial susceptibility patterns of the two isolates of E. coli
Antimicrobial agents |
E. coli E41-2 |
E. coli E41-1 |
Amikacin |
≤ 2 |
4 |
Gentamicin |
≤ 1 |
≤ 1 |
Nitrofurantoin |
≤ 16 |
≤ 16 |
Tobramycin |
≤ 1 |
2 |
Piperacillin/tazobactam |
≤ 4 |
8 |
Ampicillin/sulbactam |
≥ 32 |
≥ 32 |
Sulfamethoxazole/trimethoprim |
≥ 320 |
≥ 320 |
Imipenem |
≤ 1 |
≥ 16 |
Levofloxacin |
≥ 8 |
≥ 8 |
Ampicillin |
≥ 32 |
≥ 32 |
Aztreonam |
≥ 64 |
≥ 64 |
Ceftazidime |
≥ 64 |
≥ 64 |
Ciprofloxacin |
≥ 4 |
≥ 4 |
Ceftriaxone |
≥ 64 |
≥ 64 |
Cefotetan |
≤ 4 |
≥ 64 |
Ertapenem |
≤ 0.5 |
≥ 8 |
Cefepime |
16 |
≥ 64 |
Genome sequencing, gene annotation, and protein classification
The genome of E. coli was sequenced using a PacBio RS II platform and Illumina HiSeq X10 system (Illumina, SanDiego, CA, USA) at the Beijing Genomics Institute (Shenzhen, China). Seven databases, namely KEGG (Kyoto Encyclopedia of Genes and Genomes), COG (Clusters of Orthologous Groups), NR (Non-Redundant Protein Database), Swiss-Prot [11], GO (Gene Ontology), TrEMBL, and EggNOG were used for general function annotation. Four databases were used for pathogenicity and drug resistance analysis.
Single-nucleotide polymorphisms (SNP) analysis
Using the alignment software MUMmer (http://mummer.sourceforge.net/), each query sequence was aligned with the reference sequence. The variation sites between the query sequence and reference sequence were identified and filtered preliminarily to detect potential SNP sites. Credible SNP can be obtained by filtering the SNP located in repeat regions.
Insertion/deletion analysis
Using the LASTZ software (http://www.bx.psu.edu/miller_lab/dist/README.lastz-1.02.00/), the reference sequence and query sequence were aligned to obtain the alignment results. The alignment results were verified with BWA (http://bio-bwa.sourceforge.net/) and samtools (http://samtools.sourceforge.net/).
AMR
Both isolates of E. coli were susceptible to aminoglycoside. Following antibiotic therapy, one isolate became resistant to imipenem, ertapenem, and cefotetan. The crude enzyme of E41-1 hydrates imipenem, whereas that of E41-2 does not (Figure 1).
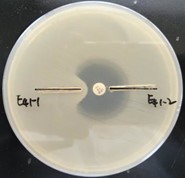
Figure 1. The hydration activity of imipenem was determined using the modified Hodge test
AMR determinants
The E41-1 complete genome (accession number: CP028483-CP028486) and E41-2 draft genome (accession number: PZPQ00000000) were submitted to the National Center for Biotechnology Information. The genome sequencing analysis showed that the carbapenem-resistant E. coli E41-1 harbored 49 types of AMR determinants. The resistance genes are listed in Table 2. Beta-lactamase gene blaCTX-M, blaTEM-1, and blaEC were associated with resistance to cephalosporin. There were 28 multidrug resistance efflux pump genes identified in the E41-1 genome. These genes encode the cell division transporter system that controls substance transport, including aminoglycoside, tigecycline, fluoroquinolone, beta-lactam, tetracycline, and fosfomycin.
Table 2. Antimicrobial resistance genes in E. coli E41-1 sequenced by whole-genome analysis
Gene_id of
E41-1 |
Identity |
Resistance
Type |
Antibiotic Resistance |
Description |
E41-1GL000027 |
99.47 |
emrd |
-- |
Multidrug resistance efflux pump. |
E41-1GL000207 |
100 |
mdtf |
doxorubicin, erythromycin |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL000208 |
100 |
mdte |
doxorubicin, erythromycin |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL000327 |
40.27 |
vanra |
vancomycin, teicoplanin |
VanA type vancomycin resistance operon genes, which can synthesize peptidoglycan with modified C-terminal D-Ala-D-Ala to D-alanine--D-lactate. |
E41-1GL000338 |
53.44 |
pbp1a |
penicillin |
The enzyme has a penicillin-insensitive transglycosylase N-terminal domain (formation of linear glycan strands) and a penicillin-sensitive transpeptidase C-terminal domain (cross-linking of the peptide subunits). |
E41-1GL000456 |
88.87 |
acrb |
aminoglycoside, glycylcycline, macrolide, beta-lactam, acriflavin |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL000457 |
68.53 |
acra |
aminoglycoside, glycylcycline, macrolide, beta-lactam, acriflavin |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL000661 |
100 |
baca |
bacitracin |
Undecaprenyl pyrophosphate phosphatase, which consists in the sequestration of Undecaprenyl pyrophosphate. |
E41-1GL000686 |
100 |
tolc |
aminoglycoside, glycylcycline, macrolide, beta-lactam, acriflavin |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL001374 |
65.95 |
acrb |
aminoglycoside, glycylcycline, macrolide, beta-lactam, acriflavin |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL001565 |
99.39 |
arna |
polymyxin |
Bifunctional enzyme that catalyzes the oxidative decarboxylation of UDP-glucuronic acid (UDP-GlcUA) to UDP-4- keto-arabinose (UDP-Ara4O) and the addition of a formyl group to UDP-4-amino-4-deoxy-L-arabinose (UDP-L- Ara4N) to form UDP-L-4-formamido-arabinose (UDP-L-Ara4FN). The modified arabinose is attached to lipid A and is required for resistance to polymyxin and cationic antimicrobial peptides. |
E41-1GL001638 |
100 |
bcr |
-- |
-- |
E41-1GL001902 |
40.28 |
vanrb |
vancomycin |
VanB type vancomycin resistance operon genes, which can synthesize peptidoglycan with modified C-terminal D-Ala-D-Ala to D-alanine--D-lactate. |
E41-1GL001931 |
100 |
emre |
aminoglycoside |
Multidrug resistance efflux pump. |
E41-1GL002260 |
100 |
mdtk |
enoxacin, norfloxacin |
Major facilitator superfamily transporter. Multidrug resistance efflux pump. |
E41-1GL002524 |
45.28 |
smec |
fluoroquinolone |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL002525 |
56.84 |
mexb |
aminoglycoside, tigecycline, fluoroquinolone, beta-lactam, tetracycline |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL002526 |
52.49 |
acra |
aminoglycoside, glycylcycline, macrolide, beta-lactam, acriflavin |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL002898 |
100 |
mdth |
deoxycholate, fosfomycin |
Major facilitator superfamily transporter. Multidrug resistance efflux pump. |
E41-1GL002911 |
100 |
mdtg |
deoxycholate, fosfomycin |
Major facilitator superfamily transporter. Multidrug resistance efflux pump. |
E41-1GL003172 |
99.69 |
macb |
macrolide |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. Macrolide-specific efflux system. |
E41-1GL003209 |
100 |
mdfa |
-- |
-- |
E41-1GL003411 |
59.9 |
pbp2 |
penicillin |
The enzyme has a penicillin-insensitive transglycosylase N-terminal domain (formation of linear glycan strands) and a penicillin-sensitive transpeptidase C-terminal domain (cross-linking of the peptide subunits). |
E41-1GL003487 |
45.26 |
oprm |
aminoglycoside, tigecycline, fluoroquinolone, beta-lactam, tetracycline |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL003551 |
76.05 |
rosa |
fosmidomycin |
Efflux pump/potassium antiporter system. RosA: Major facilitator superfamily transporter. RosB: Potassium antiporter. |
E41-1GL003552 |
80.11 |
rosb |
fosmidomycin |
Efflux pump/potassium antiporter system. RosA: Major facilitator superfamily transporter. RosB: Potassium antiporter. |
E41-1GL003567 |
100 |
acra |
aminoglycoside, glycylcycline, macrolide, beta-lactam, acriflavin |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL003568 |
99.9 |
acrb |
aminoglycoside, glycylcycline, macrolide, beta-lactam, acriflavin |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL003571 |
41.38 |
catb3 |
chloramphenicol |
Group B chloramphenicol acetyltransferase, which can inactivate chloramphenicol. Also referred to as xenobiotic acetyltransferase. |
E41-1GL003752 |
66.45 |
tet34 |
tetracycline |
Xanthine-guanine phosphoribosyl transferase. Mechanism detail unknown. |
E41-1GL003876 |
53.96 |
pbp1b |
penicillin |
The enzyme has a penicillin-insensitive transglycosylase N-terminal domain (formation of linear glycan strands) and a penicillin-sensitive transpeptidase C-terminal domain (cross-linking of the peptide subunits). |
E41-1GL003892 |
100 |
bl2be_ctxm |
monobactam, penicillin, cephalosporin_iii, ceftazidime, cephalosporin_ii, cephalosporin_i |
Class A beta-lactamase. This enzyme breaks the beta-lactam antibiotic ring open and deactivates the molecule’s antibacterial properties. |
E41-1GL003988 |
99.63 |
ksga |
kasugamycin |
Specifically, dimethylates two adjacent adenosines in the loop of a conserved hairpin near the 3'-end of 16S rRNA in the 30S particle. Its inactivation leads to kasugamycin resistance. |
E41-1GL004104 |
98.54 |
mdtm |
chloramphenicol, acriflavine, norfloxacin |
Major facilitator superfamily transporter. Multidrug resistance efflux pump. |
E41-1GL004361 |
99.47 |
bl1_ec |
cephalosporin |
Class C beta-lactamase. This enzyme breaks the beta-lactam antibiotic ring open and deactivates the molecule’s
antibacterial properties. |
E41-1GL004363 |
45.71 |
ykkc |
na_antimicrobials |
Small Multidrug Resistance (SMR) protein family. Multidrug resistance efflux pump, which consists of two proteins. |
E41-1GL004441 |
99.42 |
mdtn |
t_chloride, acriflavine, puromycin |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL004442 |
99.27 |
mdto |
t_chloride, acriflavine, puromycin |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL004443 |
99.39 |
mdtp |
t_chloride, acriflavine, puromycin |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL004853 |
99.74 |
mdtl |
chloramphenicol |
Major facilitator superfamily transporter. Multidrug resistance efflux pump. |
E41-1GL004871 |
100 |
bl2b_tem1 |
penicillin, cephalosporin_ii, cephalosporin_i |
Class A beta-lactamase. This enzyme breaks the beta-lactam antibiotic ring open and deactivates the antibacterial properties of the molecule. |
E41-1GL004876 |
100 |
teta |
tetracycline |
Major facilitator superfamily transporter, tetracycline efflux pump. |
E41-1GL004878 |
99.58 |
aph6id |
streptomycin |
Aminoglycoside O-phosphotransferase, which modifies aminoglycosides by phosphorylation. |
E41-1GL004879 |
100 |
aph33ib |
streptomycin |
Aminoglycoside O-phosphotransferase, which modifies aminoglycosides by phosphorylation. |
E41-1GL004880 |
100 |
sul2 |
sulfonamide |
Sulfonamide-resistant dihydropteroate synthase, which cannot be inhibited by sulfonamide. |
E41-1GL004890 |
100 |
sul1 |
sulfonamide |
Sulfonamide-resistant dihydropteroate synthase, which cannot be inhibited by sulfonamide. |
E41-1GL004891 |
57.09 |
ant3ia |
spectinomycin, streptomycin |
Aminoglycoside O-nucleotidylyltransferase, which modifies aminoglycosides by adenylylation. |
E41-1GL004974 |
99.73 |
tetc |
tetracycline |
Major facilitator superfamily transporter, tetracycline efflux pump. |
E41-1GL005163 |
99.45 |
qnrs |
fluoroquinolone |
Pentapeptide repeat family, which protects DNA gyrase from the inhibition of quinolones. |
Common SNP in the two isolates
There were 134 point mutations between these two isolates of E. coli. However, only 74 points were nonsynonymous mutations (Table 3). Two points of SNP were related to antibiotic resistance; one was the membrane protein E41-1GL001858 and the other was class A ESBL CTX-M-14 (E41-1GL003892). Phenotype comparison showed that these two point mutations did not alter the resistance in the two isolates. The genome chromosome and the three plasmids circos of E41-1 are shown in Figures 2-5.
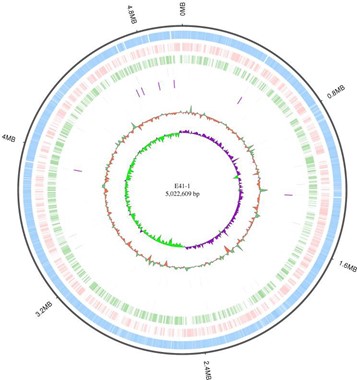
Figure 2. E41-1 genome chromosome circos
Figure 3. E41-1 plasmid plasmid 1 circos
Figure 4. E41-1 plasmid plasmid 2 circos
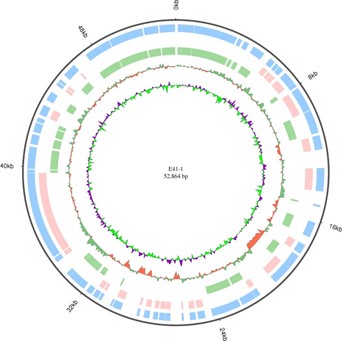
Figure 5. E41-1 plasmid plasmid 3 circos
Table 3. Nonsynonymous SNP between the two isolates of E. coli
Gene id of E41-1 |
Identity |
GenBank accession no. |
Description |
E41-1GL000108 |
99.82 |
gi|481042149|ref|WP_001296525.1| |
MULTISPECIES: L-lactate permease [Enterobacteriaceae] |
E41-1GL000133 |
99.53 |
gi|446075989|ref|WP_000153844.1| |
L-dehydroascorbate transporter large permease subunit [Escherichia coli] |
E41-1GL000358 |
99.54 |
gi|431431112|gb|ELH12890.1| |
inner membrane protein [Escherichia coli KTE165] |
E41-1GL000580 |
100 |
gi|445944916|ref|WP_000022771.1| |
MULTISPECIES: tagatose-1,6-bisphosphate aldolase [Enterobacteriaceae] |
E41-1GL000806 |
100 |
gi|291291731|gb|ADD91702.1| |
YeeS [Escherichia coli] |
E41-1GL000807 |
100 |
gi|315295288|gb|EFU54618.1| |
antirestriction protein [Escherichia coli MS 153-1] |
E41-1GL000809 |
97.4 |
gi|828391439|gb|AKK51269.1| |
yafZ [Escherichia coli PCN033] |
E41-1GL000974 |
99.77 |
gi|481041972|ref|WP_001296348.1| |
MULTISPECIES: purine permease [Enterobacteriaceae] |
E41-1GL001050 |
100 |
gi|446276418|ref|WP_000354273.1| |
VGR-related protein [Escherichia coli] |
E41-1GL001242 |
99.82 |
gi|446803683|ref|WP_000880939.1| |
MULTISPECIES: DNA repair protein RecN [Enterobacteriaceae] |
E41-1GL001548 |
99.4 |
gi|226901115|gb|EEH87374.1| |
protein yfbM [Escherichia spp. 3_2_53FAA] |
E41-1GL001636 |
99.83 |
gi|446500210|ref|WP_000578064.1| |
MULTISPECIES: ATP-dependent helicase [Enterobacteriaceae] |
E41-1GL001858 |
99.75 |
gi|446777104|ref|WP_000854360.1| |
MULTISPECIES: membrane protein [Enterobacteriaceae] |
E41-1GL002182 |
99.28 |
gi|446097161|ref|WP_000175016.1| |
MULTISPECIES: NAD(+) synthetase [Enterobacteriaceae] |
E41-1GL002283 |
99.73 |
gi|446757826|ref|WP_000835082.1| |
MULTISPECIES: anhydro-N-acetylmuramic acid kinase [Enterobacteriaceae] |
E41-1GL002307 |
99.83 |
gi|754848490|ref|WP_042209487.1| |
beta-glucuronidase [Escherichia coli] |
E41-1GL002742 |
99.61 |
gi|553359248|gb|ESA86116.1| |
Na+/H+ antiporter NhaB [Escherichia coli 907779] |
E41-1GL002798 |
99.84 |
gi|446950005|ref|WP_001027261.1| |
MULTISPECIES: Terminase large subunit from bacteriophage origin [Enterobacteriaceae] |
E41-1GL002910 |
99.68 |
gi|384470350|gb|EIE54463.1| |
lipid A biosynthesis lauroyl acyltransferase [Escherichia coli AI27] |
E41-1GL003163 |
98.4 |
gi|323958196|gb|EGB53905.1| |
AsnC family protein [Escherichia coli H263] |
E41-1GL003821 |
100 |
gi|447062911|ref|WP_001140167.1| |
MULTISPECIES: D-glycero-beta-D-manno-heptose 1,7-bisphosphate 7-phosphatase [Enterobacteriaceae] |
E41-1GL003865 |
99.79 |
gi|446768152|ref|WP_000845408.1| |
MULTISPECIES: ClC family H(+)/Cl(-) exchange transporter [Enterobacteriaceae] |
E41-1GL003892 |
100 |
gi|486436156|ref|WP_001617865.1| |
MULTISPECIES: class A extended-spectrum beta-lactamase CTX-M-14 [Enterobacteriaceae] |
E41-1GL004642 |
99.78 |
gi|446502563|ref|WP_000580417.1| |
MULTISPECIES: two-component sensor histidine kinase [Proteobacteria] |
E41-1GL005042 |
99.53 |
gi|727409755|ref|WP_033817146.1| |
protein ImpB [Escherichia coli] |
E41-1GL005057 |
100 |
gi|831357709|emb|CEL26134.1| |
ParB-like (plasmid) [Escherichia coli] |
E41-1GL005058 |
100 |
gi|446768642|ref|WP_000845898.1| |
MULTISPECIES: recombinase [Enterobacteriaceae] |
Insertion/deletion between the two isolates
Compared with E41-2, there were 12 nucleotide changes in E41-1 (nine insertion and three deletion sites). Two deletion sites and one insertion site were on the chromosome, while the other two insertion sites were located on the plasmid (Table 4). In E41-1, there was a large insertion fragment (49,097 bp) in plasmid 3 (GenBank accession no. CP028486.1) compared with E41-2. From the analysis of the coverage ratio of the E41-2 genome compared with E41-1 (Table 5), the homology of chromosome, plasmid 1, and plasmid 2 between the two isolates was very high (>99.5%). However, the coverage ratio of plasmid 3 was only 7.13%. This plasmid harbored the IMP-4 carbapenemase-coding gene and led to a change in resistance to carbapenem (Figure 6). The upstream of blaIMP-4 was the plasmid mobilization relaxosome protein Mobe gene and the downstream were class 1 integrom integrase IntI1 and IS6 family transposase IS15DIV, which may be related to carbapenemase gene capture.
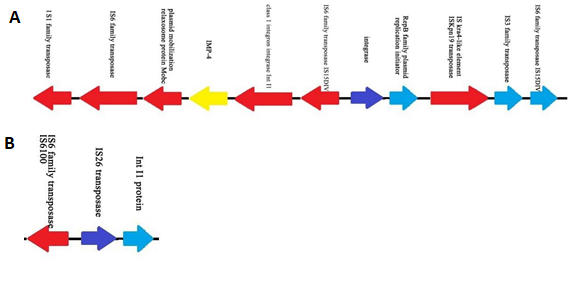
Figure 6. Structure of the E41-1 plasmid plasmid 3 (A) Features of the E41-1 plasmid plasmid 3 carrying the carbapenemase gene blaIMP-4 (B) Features of the E41-2 plasmid plasmid 3
Table 4. Indel analysis between the two isolates of E. coli
Gene ID of E41-2 |
Indel type |
InDel_start |
InDel_end |
location |
ref_start |
ref_end |
Base |
gene_ID |
Scaffold1 |
Insertion |
183631 |
183632 |
Chromosome |
2047633 |
2047633 |
C |
E41-1GL001933 |
Scaffold11 |
Insertion |
98520 |
98521 |
Chromosome |
318091 |
318091 |
G |
E41-1GL000300 |
Scaffold12 |
Deletion |
171810 |
171810 |
Chromosome |
4487177 |
4487178 |
C |
E41-1GL004379 |
Scaffold15 |
Deletion |
30739 |
30739 |
Chromosome |
2883831 |
2883832 |
G |
E41-1GL002810 |
Scaffold19 |
Deletion |
12635 |
12635 |
Chromosome |
2651515 |
2651516 |
C |
E41-1GL002552 |
Scaffold3 |
Insertion |
206329 |
206332 |
Chromosome |
209633 |
209633 |
TTA |
E41-1GL000197 |
Scaffold35 |
Insertion |
948 |
949 |
Chromosome |
833253 |
833253 |
A |
E41-1GL000819 |
Scaffold6 |
Insertion |
75123 |
75124 |
Chromosome |
3859267 |
3859267 |
T |
E41-1GL003766 |
Scaffold6 |
Insertion |
75325 |
75326 |
Chromosome |
3859385 |
3859385 |
T |
E41-1GL003766 |
Scaffold88 |
Insertion |
325 |
326 |
Plasmid2 |
20390 |
20390 |
C |
E41-1GL005107 |
Scaffold24 |
Insertion |
66053 |
66054 |
Plasmid2 |
40902 |
40902 |
C |
E41-1GL005107 |
Scaffold104 |
Insertion |
375 |
376 |
Plasmid1 |
109551 |
109551 |
G |
E41-1GL000109 |
Table 5. The coverage ratio of the E41-2 genome compared with that of E41-1
ChrID |
Reference (E41-1) size (bp) |
Covered length (bp) E41-2 |
Coverage (%) |
Chromosome |
5022609 |
5022605 |
100 |
Plasmid1 |
128911 |
128903 |
99.99 |
Plasmid2 |
86657 |
86297 |
99.58 |
Plasmid3 |
52864 |
3767 |
7.13 |
Total |
5291041 |
5241572 |
99.07 |
The rapid increase in CRE has become a global public health crisis. The resistance type and multiple acquisition pathways of CRE may affect its control in hospitals. CRE are classified into two groups: carbapenemase-producing Enterobacteriaceae (CP-CRE) and non-carbapenemase-producing Enterobacteriaceae (non-CP-CRE). Non-CP-CRE arise through mechanisms other than carbapenemase production. These mechanisms most commonly include production of ESBLs and/or AmpCs, in combination with cell membrane alterations [12]. Importantly, exposure to antibiotics may result in bacterial genomic instability that increases mutation and genetic reassortment [13]. CP-CRE has the ability to produce three different categories of carbapenemases: (a) class A (serine carbapenemases, such as Klebsiella pneumoniae carbapenemase); (b) class B (metallo-blactamases, such as IMP, VIM, and NDM); and (c) class D (OXA carbapenemases, such as OXA-23 and OXA-48) [14]. These carbapenemase-coding genes are mostly acquired through plasmids or transposons.
In our study, carbapenem-susceptible and carbapenem-resistant E. coli isolates were recovered from a single patient during therapy. Prior to the antibiotic therapy, the E. coli isolated from the sputum was susceptible to carbapenem. Following the use of carbapenem to control the infection, the E. coli became resistant to carbapenem. Based on the comparison of the genomes of the two isolates by whole-genome sequencing analysis, the sequences of chromosome, plasmid 1, and plasmid 2 of E41-1 were highly homologous to those of E41-2, and the gene difference was located in plasmid 3. Thus, we concluded that single-nucleotide mutation, insertion, and deletion did not result in the development of resistance to carbapenem. In contrast, acquisition of the IMP-4-encoding gene may be responsible for changes in drug resistance. IMP-4 carbapenemase was first identified in Acinetobacter spp. in Hong Kong between 1994 and 1998 and its encoding gene was not associated with any plasmids [15]. In 2001, the occurrence of the blaIMP-4 gene on a conjugative plasmid in Citrobacter youngae was reported by Hawkey et al. [16]. Based on the results of our study, the blaIMP-4 gene was related to class 1 integron IntI1, IS6 family transpose, and plasmid mobilization relaxosome protein Mobe gene, which may have led to gene capture.
In recent years, WGS has been widely used in genomic research. It provides information on the arrangement of multiple AMR genes and associated genes. Because gene expression is influenced by numerous factors, detection of an AMR gene does not necessarily indicate that the isolate is resistant to some antibiotic. However, WGS is more sensitive and comprehensive than traditional methods and facilitates the identification of more determinants [17]. A total of 49 types of AMR genes were determined in the chromosome of E41-1. Of those, 30 AMR genes exhibited highly homologous identities to those of previously confirmed AMR genes (>99%). Most of the genes (acra, acrb, tolc, smec, mexb, emre, et cl) were efflux pump genes associated with multi-antibiotics, including aminoglycoside, tigecycline, fluoroquinolone, beta-lactam, tetracycline, and fosfomycin. In contrast, E41-1 was susceptible to aminoglycoside. Hence, even if the isolate harbors the emre gene, it may not be resistant to aminoglycoside. The phenotype of the isolate is not determined by a single AMR gene, but is affected by numerous factors. Unlike traditional biocuration, the AMR genes are constantly moving and mutating under selective pressures. New AMR mutations and horizontal transfer of AMR genes among pathogens will occur with the emergence of new threats from the environment and protoresistome [10,18,19]. We should develop proper analytical pipelines for the accurate detection of the resistome and subsequent accurate prediction of the antibiogram based on genomic and metagenomic data [20]. Thus, WGS can be employed inmodifying antibiotic usage strategies to optimize antimicrobial stewardship.
In principle, for the clinical prediction of antibiotic resistance by WGS, a comprehensive set of genetic determinants of the resistome needs to be identified for each species and further research should investigate new mechanisms of antimicrobial resistance. Recent studies demonstrated that WGS could be feasibly and effectively used for surveillance of antibiotic resistance and provides actionable results in infection control.
Whole-genome sequencing was applied to analyze the genetic difference between the two isolates of Escherichia coli. We found that single-nucleotide mutation did not contribute to change in antibiotic resistance. Acquisition of a plasmid carrying a migratable gene cassette resulted in spreading of antibiotic resistance.
This study was supported by the National Natural Science Foundation of China (81101282).
Qiong Wu and Jun Hu contributed equally to this work. Qiong Wu and Jun Hu conceived and designed research. Jianqiang Wang, Yungai Li and Yunqi Pan conducted experiments. Rong Chen and Jin Tang analyzed data. Qiong Wu and Jin Tang wrote the manuscript. All authors read and approved the manuscript.
- Xu Y, Gu B, Huang M, Liu H, Xu T, et al. (2015) Epidemiology of carbapenem resistant Enterobacteriaceae (CRE) during 2000-2012 in Asia. J Thorac Dis 7: 376-385. [Crossref]
- Iredell J, Brown J, Tagg K (2016) Antibiotic resistance in Enterobacteriaceae: mechanisms and clinical implications. BMJ 352: h6420. [Crossref]
- Hu FP, Guo Y, Zhu DM, Wang F, Jiang XF, et al. (2016) Resistance trends among clinical isolates in China reported from CHINET surveillance of bacterial resistance, 2005-2014. Clin Microbiol Infect 22: S9-S14. [Crossref]
- Livingstone D, Gill MJ, Wise R (1995) Mechanisms of resistance to the carbapenems. J Antimicrobial Chemother 35: 1-5.
- Goodman KE, Simner PJ, Tamma PD, Milstone AM (2016) Infection control implications of heterogeneous resistance mechanisms in carbapenem-resistant Enterobacteriaceae (CRE). Expert Rev Anti Infect Ther 14: 95-108. [Crossref]
- Yu H, Qu F, Shan B, Huang B, Jia W, et al. (2016) Detection of the mcr-1 Colistin Resistance Gene in Carbapenem-Resistant Enterobacteriaceae from Different Hospitals in China. Antimicrob Agents Chemother 60: 5033-5035. [Crossref]
- Snitkin ES, Zelazny AM, Thomas PJ, Stock F; NISC Comparative Sequencing Program Group, et al. (2012) Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med 4: 148ra116. [Crossref]
- Onori R, Gaiarsa S, Comandatore F, Pongolini S, Brisse S, et al. (2015) Tracking Nosocomial Klebsiella pneumoniae Infections and Outbreaks by Whole-Genome Analysis: Small-Scale Italian Scenario within a Single Hospital. J Clin Microbiol 53: 2861-2868. [Crossref]
- Knudsen PK, Gammelsrud KW, Alfsnes K, Steinbakk M, Abrahamsen TG, et al. (2018) Transfer of a bla CTX-M-1-carrying plasmid between different Escherichia coli strains within the human gut explored by whole genome sequencing analyses. Sci Rep 8: 280. [Crossref]
- Gillings MR, Paulsen IT, Tetu SG (2017) Genomics and the evolution of antibiotic resistance. Ann N Y Acad Sci 1388: 92-107. [Crossref]
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M (2016) KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44: D457-462.
- Rock C, Thom KA, Masnick M, Johnson JK, Harris AD, et al. (2014) Frequency of Klebsiella pneumoniae carbapenemase (KPC)-producing and non-KPC-producing Klebsiella species contamination of healthcare workers and the environment. Infect Control Hosp Epidemiol 35: 426-429. [Crossref]
- Shapiro RS (2015) Antimicrobial-induced DNA damage and genomic instability in microbial pathogens. PLoS Pathog 11: e1004678. [Crossref]
- AlTamimi M, AlSalamah A, AlKhulaifi M, AlAjlan H (2017) Comparison of phenotypic and PCR methods for detection of carbapenemases production by Enterobacteriaceae. Saudi J Biol Sci 24: 155-161. [Crossref]
- Riccio ML, Franceschini N, Boschi L, Caravelli B, Cornaglia G, et al. (2000) Characterization of the metallo-beta-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of bla(IMP) allelic variants carried by gene cassettes of different phylogeny. Antimicrob Agents Chemother 44: 1229-1235. [Crossref]
- Hawkey PM, Xiong J, Ye H, Li H, M'Zali FH (2001) Occurrence of a new metallo-beta-lactamase IMP-4 carried on a conjugative plasmid in Citrobacter youngae from the People's Republic of China. FEMS Microbiol Lett 194: 53-57. [Crossref]
- Su JQ, Cui L, Chen QL, An XL, Zhu YG (2017) Application of genomic technologies to measure and monitor antibiotic resistance in animals. Ann N Y Acad Sci 1388: 121-135. [Crossref]
- Köser CU, Ellington MJ, Peacock SJ (2014) Whole-genome sequencing to control antimicrobial resistance. Trends Genet 30: 401-407. [Crossref]
- Schürch AC, van Schaik W (2017) Challenges and opportunities for whole-genome sequencing-based surveillance of antibiotic resistance. Ann N Y Acad Sci 1388: 108-120. [Crossref]
- McArthur AG, Tsang KK (2017) Antimicrobial resistance surveillance in the genomic age. Ann N Y Acad Sci 1388: 78-91. [Crossref]
Editorial Information
Editor-in-Chief
Yeun-Hwa Gu
Junshin Gakuen University, Japan
Article type
Research Article
Publication History
Received: April 12, 2021
Accepted: May 17, 2021
Published: May 25, 2021
Copyright
©2021 Wu Q. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation
Wu Q, Wang J, Li Y, Pan Y, Tang J (2021) Genomic differences between carbapenem-susceptible and carbapenem-resistant Escherichia coli analyzed by whole-genome sequencing. Clin Microbiol Infect Dis 6: DOI: 10.15761/CMID.1000187.

Figure 1. The hydration activity of imipenem was determined using the modified Hodge test

Figure 2. E41-1 genome chromosome circos

Figure 3. E41-1 plasmid plasmid 1 circos

Figure 4. E41-1 plasmid plasmid 2 circos

Figure 5. E41-1 plasmid plasmid 3 circos

Figure 6. Structure of the E41-1 plasmid plasmid 3 (A) Features of the E41-1 plasmid plasmid 3 carrying the carbapenemase gene blaIMP-4 (B) Features of the E41-2 plasmid plasmid 3
Table 1. Antimicrobial susceptibility patterns of the two isolates of E. coli
Antimicrobial agents |
E. coli E41-2 |
E. coli E41-1 |
Amikacin |
≤ 2 |
4 |
Gentamicin |
≤ 1 |
≤ 1 |
Nitrofurantoin |
≤ 16 |
≤ 16 |
Tobramycin |
≤ 1 |
2 |
Piperacillin/tazobactam |
≤ 4 |
8 |
Ampicillin/sulbactam |
≥ 32 |
≥ 32 |
Sulfamethoxazole/trimethoprim |
≥ 320 |
≥ 320 |
Imipenem |
≤ 1 |
≥ 16 |
Levofloxacin |
≥ 8 |
≥ 8 |
Ampicillin |
≥ 32 |
≥ 32 |
Aztreonam |
≥ 64 |
≥ 64 |
Ceftazidime |
≥ 64 |
≥ 64 |
Ciprofloxacin |
≥ 4 |
≥ 4 |
Ceftriaxone |
≥ 64 |
≥ 64 |
Cefotetan |
≤ 4 |
≥ 64 |
Ertapenem |
≤ 0.5 |
≥ 8 |
Cefepime |
16 |
≥ 64 |
Table 2. Antimicrobial resistance genes in E. coli E41-1 sequenced by whole-genome analysis
Gene_id of
E41-1 |
Identity |
Resistance
Type |
Antibiotic Resistance |
Description |
E41-1GL000027 |
99.47 |
emrd |
-- |
Multidrug resistance efflux pump. |
E41-1GL000207 |
100 |
mdtf |
doxorubicin, erythromycin |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL000208 |
100 |
mdte |
doxorubicin, erythromycin |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL000327 |
40.27 |
vanra |
vancomycin, teicoplanin |
VanA type vancomycin resistance operon genes, which can synthesize peptidoglycan with modified C-terminal D-Ala-D-Ala to D-alanine--D-lactate. |
E41-1GL000338 |
53.44 |
pbp1a |
penicillin |
The enzyme has a penicillin-insensitive transglycosylase N-terminal domain (formation of linear glycan strands) and a penicillin-sensitive transpeptidase C-terminal domain (cross-linking of the peptide subunits). |
E41-1GL000456 |
88.87 |
acrb |
aminoglycoside, glycylcycline, macrolide, beta-lactam, acriflavin |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL000457 |
68.53 |
acra |
aminoglycoside, glycylcycline, macrolide, beta-lactam, acriflavin |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL000661 |
100 |
baca |
bacitracin |
Undecaprenyl pyrophosphate phosphatase, which consists in the sequestration of Undecaprenyl pyrophosphate. |
E41-1GL000686 |
100 |
tolc |
aminoglycoside, glycylcycline, macrolide, beta-lactam, acriflavin |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL001374 |
65.95 |
acrb |
aminoglycoside, glycylcycline, macrolide, beta-lactam, acriflavin |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL001565 |
99.39 |
arna |
polymyxin |
Bifunctional enzyme that catalyzes the oxidative decarboxylation of UDP-glucuronic acid (UDP-GlcUA) to UDP-4- keto-arabinose (UDP-Ara4O) and the addition of a formyl group to UDP-4-amino-4-deoxy-L-arabinose (UDP-L- Ara4N) to form UDP-L-4-formamido-arabinose (UDP-L-Ara4FN). The modified arabinose is attached to lipid A and is required for resistance to polymyxin and cationic antimicrobial peptides. |
E41-1GL001638 |
100 |
bcr |
-- |
-- |
E41-1GL001902 |
40.28 |
vanrb |
vancomycin |
VanB type vancomycin resistance operon genes, which can synthesize peptidoglycan with modified C-terminal D-Ala-D-Ala to D-alanine--D-lactate. |
E41-1GL001931 |
100 |
emre |
aminoglycoside |
Multidrug resistance efflux pump. |
E41-1GL002260 |
100 |
mdtk |
enoxacin, norfloxacin |
Major facilitator superfamily transporter. Multidrug resistance efflux pump. |
E41-1GL002524 |
45.28 |
smec |
fluoroquinolone |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL002525 |
56.84 |
mexb |
aminoglycoside, tigecycline, fluoroquinolone, beta-lactam, tetracycline |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL002526 |
52.49 |
acra |
aminoglycoside, glycylcycline, macrolide, beta-lactam, acriflavin |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL002898 |
100 |
mdth |
deoxycholate, fosfomycin |
Major facilitator superfamily transporter. Multidrug resistance efflux pump. |
E41-1GL002911 |
100 |
mdtg |
deoxycholate, fosfomycin |
Major facilitator superfamily transporter. Multidrug resistance efflux pump. |
E41-1GL003172 |
99.69 |
macb |
macrolide |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. Macrolide-specific efflux system. |
E41-1GL003209 |
100 |
mdfa |
-- |
-- |
E41-1GL003411 |
59.9 |
pbp2 |
penicillin |
The enzyme has a penicillin-insensitive transglycosylase N-terminal domain (formation of linear glycan strands) and a penicillin-sensitive transpeptidase C-terminal domain (cross-linking of the peptide subunits). |
E41-1GL003487 |
45.26 |
oprm |
aminoglycoside, tigecycline, fluoroquinolone, beta-lactam, tetracycline |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL003551 |
76.05 |
rosa |
fosmidomycin |
Efflux pump/potassium antiporter system. RosA: Major facilitator superfamily transporter. RosB: Potassium antiporter. |
E41-1GL003552 |
80.11 |
rosb |
fosmidomycin |
Efflux pump/potassium antiporter system. RosA: Major facilitator superfamily transporter. RosB: Potassium antiporter. |
E41-1GL003567 |
100 |
acra |
aminoglycoside, glycylcycline, macrolide, beta-lactam, acriflavin |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL003568 |
99.9 |
acrb |
aminoglycoside, glycylcycline, macrolide, beta-lactam, acriflavin |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL003571 |
41.38 |
catb3 |
chloramphenicol |
Group B chloramphenicol acetyltransferase, which can inactivate chloramphenicol. Also referred to as xenobiotic acetyltransferase. |
E41-1GL003752 |
66.45 |
tet34 |
tetracycline |
Xanthine-guanine phosphoribosyl transferase. Mechanism detail unknown. |
E41-1GL003876 |
53.96 |
pbp1b |
penicillin |
The enzyme has a penicillin-insensitive transglycosylase N-terminal domain (formation of linear glycan strands) and a penicillin-sensitive transpeptidase C-terminal domain (cross-linking of the peptide subunits). |
E41-1GL003892 |
100 |
bl2be_ctxm |
monobactam, penicillin, cephalosporin_iii, ceftazidime, cephalosporin_ii, cephalosporin_i |
Class A beta-lactamase. This enzyme breaks the beta-lactam antibiotic ring open and deactivates the molecule’s antibacterial properties. |
E41-1GL003988 |
99.63 |
ksga |
kasugamycin |
Specifically, dimethylates two adjacent adenosines in the loop of a conserved hairpin near the 3'-end of 16S rRNA in the 30S particle. Its inactivation leads to kasugamycin resistance. |
E41-1GL004104 |
98.54 |
mdtm |
chloramphenicol, acriflavine, norfloxacin |
Major facilitator superfamily transporter. Multidrug resistance efflux pump. |
E41-1GL004361 |
99.47 |
bl1_ec |
cephalosporin |
Class C beta-lactamase. This enzyme breaks the beta-lactam antibiotic ring open and deactivates the molecule’s
antibacterial properties. |
E41-1GL004363 |
45.71 |
ykkc |
na_antimicrobials |
Small Multidrug Resistance (SMR) protein family. Multidrug resistance efflux pump, which consists of two proteins. |
E41-1GL004441 |
99.42 |
mdtn |
t_chloride, acriflavine, puromycin |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL004442 |
99.27 |
mdto |
t_chloride, acriflavine, puromycin |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL004443 |
99.39 |
mdtp |
t_chloride, acriflavine, puromycin |
Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. |
E41-1GL004853 |
99.74 |
mdtl |
chloramphenicol |
Major facilitator superfamily transporter. Multidrug resistance efflux pump. |
E41-1GL004871 |
100 |
bl2b_tem1 |
penicillin, cephalosporin_ii, cephalosporin_i |
Class A beta-lactamase. This enzyme breaks the beta-lactam antibiotic ring open and deactivates the antibacterial properties of the molecule. |
E41-1GL004876 |
100 |
teta |
tetracycline |
Major facilitator superfamily transporter, tetracycline efflux pump. |
E41-1GL004878 |
99.58 |
aph6id |
streptomycin |
Aminoglycoside O-phosphotransferase, which modifies aminoglycosides by phosphorylation. |
E41-1GL004879 |
100 |
aph33ib |
streptomycin |
Aminoglycoside O-phosphotransferase, which modifies aminoglycosides by phosphorylation. |
E41-1GL004880 |
100 |
sul2 |
sulfonamide |
Sulfonamide-resistant dihydropteroate synthase, which cannot be inhibited by sulfonamide. |
E41-1GL004890 |
100 |
sul1 |
sulfonamide |
Sulfonamide-resistant dihydropteroate synthase, which cannot be inhibited by sulfonamide. |
E41-1GL004891 |
57.09 |
ant3ia |
spectinomycin, streptomycin |
Aminoglycoside O-nucleotidylyltransferase, which modifies aminoglycosides by adenylylation. |
E41-1GL004974 |
99.73 |
tetc |
tetracycline |
Major facilitator superfamily transporter, tetracycline efflux pump. |
E41-1GL005163 |
99.45 |
qnrs |
fluoroquinolone |
Pentapeptide repeat family, which protects DNA gyrase from the inhibition of quinolones. |
Table 3. Nonsynonymous SNP between the two isolates of E. coli
Gene id of E41-1 |
Identity |
GenBank accession no. |
Description |
E41-1GL000108 |
99.82 |
gi|481042149|ref|WP_001296525.1| |
MULTISPECIES: L-lactate permease [Enterobacteriaceae] |
E41-1GL000133 |
99.53 |
gi|446075989|ref|WP_000153844.1| |
L-dehydroascorbate transporter large permease subunit [Escherichia coli] |
E41-1GL000358 |
99.54 |
gi|431431112|gb|ELH12890.1| |
inner membrane protein [Escherichia coli KTE165] |
E41-1GL000580 |
100 |
gi|445944916|ref|WP_000022771.1| |
MULTISPECIES: tagatose-1,6-bisphosphate aldolase [Enterobacteriaceae] |
E41-1GL000806 |
100 |
gi|291291731|gb|ADD91702.1| |
YeeS [Escherichia coli] |
E41-1GL000807 |
100 |
gi|315295288|gb|EFU54618.1| |
antirestriction protein [Escherichia coli MS 153-1] |
E41-1GL000809 |
97.4 |
gi|828391439|gb|AKK51269.1| |
yafZ [Escherichia coli PCN033] |
E41-1GL000974 |
99.77 |
gi|481041972|ref|WP_001296348.1| |
MULTISPECIES: purine permease [Enterobacteriaceae] |
E41-1GL001050 |
100 |
gi|446276418|ref|WP_000354273.1| |
VGR-related protein [Escherichia coli] |
E41-1GL001242 |
99.82 |
gi|446803683|ref|WP_000880939.1| |
MULTISPECIES: DNA repair protein RecN [Enterobacteriaceae] |
E41-1GL001548 |
99.4 |
gi|226901115|gb|EEH87374.1| |
protein yfbM [Escherichia spp. 3_2_53FAA] |
E41-1GL001636 |
99.83 |
gi|446500210|ref|WP_000578064.1| |
MULTISPECIES: ATP-dependent helicase [Enterobacteriaceae] |
E41-1GL001858 |
99.75 |
gi|446777104|ref|WP_000854360.1| |
MULTISPECIES: membrane protein [Enterobacteriaceae] |
E41-1GL002182 |
99.28 |
gi|446097161|ref|WP_000175016.1| |
MULTISPECIES: NAD(+) synthetase [Enterobacteriaceae] |
E41-1GL002283 |
99.73 |
gi|446757826|ref|WP_000835082.1| |
MULTISPECIES: anhydro-N-acetylmuramic acid kinase [Enterobacteriaceae] |
E41-1GL002307 |
99.83 |
gi|754848490|ref|WP_042209487.1| |
beta-glucuronidase [Escherichia coli] |
E41-1GL002742 |
99.61 |
gi|553359248|gb|ESA86116.1| |
Na+/H+ antiporter NhaB [Escherichia coli 907779] |
E41-1GL002798 |
99.84 |
gi|446950005|ref|WP_001027261.1| |
MULTISPECIES: Terminase large subunit from bacteriophage origin [Enterobacteriaceae] |
E41-1GL002910 |
99.68 |
gi|384470350|gb|EIE54463.1| |
lipid A biosynthesis lauroyl acyltransferase [Escherichia coli AI27] |
E41-1GL003163 |
98.4 |
gi|323958196|gb|EGB53905.1| |
AsnC family protein [Escherichia coli H263] |
E41-1GL003821 |
100 |
gi|447062911|ref|WP_001140167.1| |
MULTISPECIES: D-glycero-beta-D-manno-heptose 1,7-bisphosphate 7-phosphatase [Enterobacteriaceae] |
E41-1GL003865 |
99.79 |
gi|446768152|ref|WP_000845408.1| |
MULTISPECIES: ClC family H(+)/Cl(-) exchange transporter [Enterobacteriaceae] |
E41-1GL003892 |
100 |
gi|486436156|ref|WP_001617865.1| |
MULTISPECIES: class A extended-spectrum beta-lactamase CTX-M-14 [Enterobacteriaceae] |
E41-1GL004642 |
99.78 |
gi|446502563|ref|WP_000580417.1| |
MULTISPECIES: two-component sensor histidine kinase [Proteobacteria] |
E41-1GL005042 |
99.53 |
gi|727409755|ref|WP_033817146.1| |
protein ImpB [Escherichia coli] |
E41-1GL005057 |
100 |
gi|831357709|emb|CEL26134.1| |
ParB-like (plasmid) [Escherichia coli] |
E41-1GL005058 |
100 |
gi|446768642|ref|WP_000845898.1| |
MULTISPECIES: recombinase [Enterobacteriaceae] |
Table 4. Indel analysis between the two isolates of E. coli
Gene ID of E41-2 |
Indel type |
InDel_start |
InDel_end |
location |
ref_start |
ref_end |
Base |
gene_ID |
Scaffold1 |
Insertion |
183631 |
183632 |
Chromosome |
2047633 |
2047633 |
C |
E41-1GL001933 |
Scaffold11 |
Insertion |
98520 |
98521 |
Chromosome |
318091 |
318091 |
G |
E41-1GL000300 |
Scaffold12 |
Deletion |
171810 |
171810 |
Chromosome |
4487177 |
4487178 |
C |
E41-1GL004379 |
Scaffold15 |
Deletion |
30739 |
30739 |
Chromosome |
2883831 |
2883832 |
G |
E41-1GL002810 |
Scaffold19 |
Deletion |
12635 |
12635 |
Chromosome |
2651515 |
2651516 |
C |
E41-1GL002552 |
Scaffold3 |
Insertion |
206329 |
206332 |
Chromosome |
209633 |
209633 |
TTA |
E41-1GL000197 |
Scaffold35 |
Insertion |
948 |
949 |
Chromosome |
833253 |
833253 |
A |
E41-1GL000819 |
Scaffold6 |
Insertion |
75123 |
75124 |
Chromosome |
3859267 |
3859267 |
T |
E41-1GL003766 |
Scaffold6 |
Insertion |
75325 |
75326 |
Chromosome |
3859385 |
3859385 |
T |
E41-1GL003766 |
Scaffold88 |
Insertion |
325 |
326 |
Plasmid2 |
20390 |
20390 |
C |
E41-1GL005107 |
Scaffold24 |
Insertion |
66053 |
66054 |
Plasmid2 |
40902 |
40902 |
C |
E41-1GL005107 |
Scaffold104 |
Insertion |
375 |
376 |
Plasmid1 |
109551 |
109551 |
G |
E41-1GL000109 |
Table 5. The coverage ratio of the E41-2 genome compared with that of E41-1
ChrID |
Reference (E41-1) size (bp) |
Covered length (bp) E41-2 |
Coverage (%) |
Chromosome |
5022609 |
5022605 |
100 |
Plasmid1 |
128911 |
128903 |
99.99 |
Plasmid2 |
86657 |
86297 |
99.58 |
Plasmid3 |
52864 |
3767 |
7.13 |
Total |
5291041 |
5241572 |
99.07 |






