Introduction: Since the emergence of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), real-time reverse-transcription polymerase chain reaction (RT-PCR)-based testing remains the gold standard for its laboratory diagnosis. Due to the increasing diagnosis demand, resources for RT-PCR testing are limited in many countries, especially in Africa. Here, we evaluated the LumiraDx SARS-CoV-2 antigen test for alternative diagnosis of SARS-CoV-2 in hospital setting.
Materials and methods: This prospective validation study was conducted among 184 subjected presented at the hospital for COVID-19 testing. Nasal swap samples have been collected into 0.7 mL of the extraction buffer for LumiraDx antigen test. In parallel, oropharyngeal and nasopharyngeal sap samples were collected into viral transport media for RT-PCR testing.
Results: Compared to the RT-PCR, the sensitivity and specificity of the LumiraDx antigen SARS-CoV-2 test were 81.6% [95%CI 66.8-91.3] and 100% [95%CI 96.4-100] respectively. Given to the threshold cycle (Ct)the sensitivity and specificity were 94.4% [95%CI 79.9-99.0] and 100% [95%CI 96.4-100], respectively when the Ct value was below or equal 33 cycles, and 27.2% [95%CI 7.3-60.6] and 100% [95%CI 96.4-100] when it was above 33. The kappa coefficient showed was 0.86 when considering all the patients and 0.96 for Ct values below 30 cycles. The agreement with RT-PCR ranged from was 88.2 to 100% in patients within 12 days of the onset of symptoms.
Conclusions: Our data have shown that the LumiraDx antigen SARS-Co-2 test elicits high sensitivity and good agreement when compared to the RT-PCR and can significantly improve the SARS-CoV-2 diagnosis in clinic-based settings.
LumiraDx, SARS-CoV-2, PCR, clinical setting
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that emerged in Wuhan, China, rapidly spread across the world [1,2] and has been declared as a major public health emergency worldwide [3]. Because of continuous progression despite the global effort, rapid and accurate identification of infected subjects for their isolation are required for better control the pandemic. Since its emergence, real-time reverse transcription polymerase chain reaction (RT-PCR) has been considered the gold standard of SARS-CoV-2 diagnostic as recommended by the World Health Organization (WHO) and the Centers for Disease Control (CDC) [4,5]. However, like other molecular methods such as loop-mediated isothermal amplification-based assay (RT-LAMP), microarray, and high-throughput sequencing, RT-PCR appears costly and requires special equipment and skilled laboratory personnel. Consequently, shortages of PCR kits and related consumables because of the increasing worldwide demand are major constraints for molecular methods. Moreover, PCR testing provides SARS-CoV2 test results in much longer time, which may delay the ability people to stay isolated to prevent transmission and the spread of the virus [6,7].
The development of point-of-care tests (POCTs) as alternative assay for SARS-CoV-2 infection diagnosis is of higher importance, especially in Africa where most of the clinical laboratories have limited capacities. When used alone or as a complement to PCR-based testing, such affordable easy-to-use emerging diagnostic tools have the potential to play a significant role in guiding patient management, public health interventions, and disease surveillance. Numerous commercial antigen diagnostic assays are now available [8-17] and studies have shown sensitivity of SARS-CoV-2 Ag ranging between 22.9% and 93.9% compared to RT-PCR [9,12-14,18].
Most of the time, the SARS-CoV-2 antigen tests are based on the lateral flow principle and work with respiratory specimens such as oral, nasal or nasopharyngeal. Nevertheless, other immunofluorescence-based POC antigen tests, like the LumiraDx SARS-CoV-2 platform are also available in the market [19]. The LumiraDx SARS-CoV-2 antigen test (LumiraDx UK Ltd., Dumyat Business Park, Alloa, FK10 2PB, UK) is a microfluidic immunofluorescence assay for the direct and qualitative detection of nucleocapsid protein. It runs on a portable, wall outlet or battery-powered multi-assay point-of-care instrument [20,21]. The assay reagents are dry single-use, disposable, microfluidic test strips that contain specific antibodies to form an immunoassay complex that uses a fluorescent latex signal to detect the N protein of SARS-CoV-2 in a test sample [22]. Like numerus emerging antigen tests that obtained emergency authorization use, there are limited data on their performance. The objective of this study was to evaluate the clinical performance of the LumiraDx™ Platform for the identification of SARS-COV-2 in clinical setting compared to RT-PCR.
Study populations and samples
A prospective validation study of the LumiraDx SARS-CoV-2 Ag test was conducted among 184 who presented at the Institute of Social Hygiene (Dakar, Senegal) for COVID-19 testing. Operators have been trained in collecting and testing specimens using the LumiraDx device. Nasal swaps provided with the kit (SteriPack™ Sterile Polyester Spun Swab, product code: 60564REVA) have been used for collection of nasal swap specimens in both nostrils. The swap was then placed into 0.7 ml of the extraction buffer for LumiraDx SARS-CoV-2 antigen test. The oropharyngeal and nasopharyngeal samples were also collected into 2 ml of viral transport media (KANG JIAN Virus Preservation Medium (Cat: 0201101)) for RT-PCR testing. Questionnaire reporting clinical characteristics have been used for each participant. This study was approved by the “Comité National d’Ethique pour la recherche en Santé” of the Senegalese Ministry of Health (Authorisation Nr: 00000110/MSAS/CNERS/Sec).
Real-time reverse-transcription polymerase chain reaction testing
RNA extraction: The oropharyngeal and nasopharyngeal samples were first inactivated in a water bath at 90°C for 30 min. The samples were then aliquoted in 1.5 ml vial and RNA was extracted with MagMAX Viral/Pathogen II Nucleic Acid isolation kit using the Kingfisher platform according to the guidelines of manufacturing and eluted in 50 μL (Thermo Fisher Scientific, Waltham, MA USA, www.thermofisher.com).
Real-time Reverse transcriptase-polymerase chain reaction: RNA elution plates were stored at 4°C while preparing master mix. For SARS-CoV-2 detection, Allplex™ 2019-nCoV assay from Seegene Inc. were used according to the manufacturer protocol. Briefly, a master mix of 5μl of 2019-nCoV MOM, 5μl of buffer 5X, 5μl of RNase-free water, 1μl of internal control (IC) and 2μl of enzymes per sample including negative and positive control were mixed. In each well, 18μl of master mix were distributed and either 8μl of sample added, 8μl of positive control or 8μl of RNase-free water for negative control. Plates were then spun down at 2500 rpm for 5 s and analysed on a CFX96 Touch Real-Time PCR from BioRad. Reverse Transcription reaction 1 cycle: 50°C/20 min – 95°C/15 min. PCR reaction 45 cycles: 94°C/15 s – 60°C/30 sec- 72°C/15 sec. Fluorescence was measured at 60°C and 72°C using channels FAM (E gene), HEX (IC), Cal Red 610 (RdRP) and Quasar 670 (N gene). Results were compiled and analyzed using 2019-nCoV viewer from Seegene Inc. according to the manufacturer’s instructions (Seegene. AllplexTM 2019-nCoV Assay, Cat no. RP10250X / RP10252W) [23]. Results were defined as positive if the viral genome was detected at threshold cycle (Ct) values ≤ 35, as indeterminate at Ct values >35 and ≤ 38, and as negative at Ct values >38 with internal control positive.
LumiraDx SARS-CoV-2 antigen test processing
For each strip lot, all the LumiraDx devices underwent lot calibration files as recommended by the manufacturer, to provide the instruments with information needed to perform diagnostic tests. After nasal swab collection, sample extraction was performed by introducing the patient’s swap into the extraction vial containing 0.7 ml of extraction buffer (reference Nr: Spec-32384 Rev1) by soaking for 10 seconds and stirring well by rotating the swab against the side of the vial about 5 times. The patient swab was then removed while squeezing the middle of the extraction vial. After inverting gently five times, a drop of the extracted sample was applied onto the sample application area of the inserted test strip (reference Nr: Spec-32762). When the sample was detected, the test took 12 min to deliver a positive or negative result. The instrument platform was connected to a cloud server for uploading test data into electronic medical records. All buffer specimens for nasopharyngeal swabs were tested fresh at the clinical site within 1 h of collection.
Statistical analysis
To evaluate the diagnosis performance, results have been stratified by gender, age, days since symptom the first symptom was reported, and the RT-PCR cycle threshold. Sensitivity, specificity, positive likelihood ratios, and 95% CI were determined. Inter-Rater Reliability was estimated in SPSS 20 using Cohen's kappa which corrects agreement due by chance.
Participant characteristics
From March to April 2021, a total of 184 nasal swap samples were collected from individuals presented at the hospital for COVD-19 testing for LumiraDx antigen SARS-CoV-2 test. Parallelly, oral and nasopharyngeal swab samples were collected from the same subjects for RT-PCR testing. Participants were aged from 16 to 96 years, including 106 female (57.1%) and 78 males (42.9%). 172 (93.4%) participants presented COVID-19-related symptoms for which the duration when from 0 to 40 days.
Clinical performance of the SARS-CoV-2 Antigen assay
Among 184 subjects, 44 nasopharyngeal swab samples were positive for SARS-CoV-2 using the RT-PCR testing results, giving an overall estimated prevalence of 23.9% in this cohort for the molecular method. In nasal swap samples, LumiraDx SARS-CoV-2 antigen test elicited 34 positive samples corresponding to a prevalence of 19.7%. Within the 44 RT-PCR-positive subjects, le LumiraDx detected 36 positive individuals (81.8%) (Table 1).
Table 1. Inter Reliability Rate (IRR) of all subjects, positive samples and the symptomatic individuals by day of day the symptom onset. IRRs were assessed using matched from RT-PCR and LumiraDx results, and total results population tested. Statistical analysis was performed using SPSS version 20 (IBM, Inc.)
Day of symptoms |
|
All symptomatic |
Positive |
D0 |
D1 |
D2 |
D3 |
D4 |
D5 |
D6 |
D7 |
D8 |
D9 |
D10 |
D11 |
D12 |
> 12 days |
Match |
172 |
32 |
8 |
12 |
14 |
15 |
15 |
12 |
12 |
11 |
8 |
13 |
15 |
9 |
11 |
19 |
Total |
183 |
44 |
8 |
12 |
15 |
17 |
16 |
12 |
13 |
11 |
9 |
13 |
15 |
9 |
12 |
21 |
IRR |
94.0% |
72.7% |
100% |
100% |
93.3% |
88.2% |
93.80% |
100% |
92.3% |
100% |
88.9% |
100% |
100% |
100% |
91.7% |
85.7% |
When compared to RT-PCR, the LumiraDx SARS-CoV-2 antigen assay displayed an overall sensitivity of 81.8% [95% CI 66.8-91.3%] and specificity of 100% [95% CI 96.4-100%] for nasal swab samples, when considering all the participants symptoms (Table 1). Analysis according to the threshold Ct value has shown sensibility and specificity of 94.4% [95% CI 79.9-99.0%] and 100% [95% CI 96.6-100%] respectively when Ct values are below or equal 33 cycles; above 33 cycles, the sensitivity and specificity were 27.2% [95% CI 7.3-60.6] and 100% [95% CI 96.6-100] respectively (Table 1). Analysis of the Cohen’s kappa coefficient showed a good overall agreement between the RT-PRC and LumiraDx antigen with k=0.86. Reliably of both methods increase when for Ct values below and equal 33 cycles with k=0.96 while it was for Ct values were above 33 cycles (k=0.41) (Table 1).
When analysing the agreement between RT-PCR and LumiraDx with symptoms, this was 94% when considering all the symptomatic subjects. Within 12 days of the onset of symptoms, the LumiraDx test elicited an agreement with RT-PCR from 88.2% to 100%, and 85.7% when symptoms appear after 12 days (Table 2). When plotting the Ct values against the time the symptom onset, we found a trend of higher median of Ct values from day 5 the symptom onset (Figure 1).
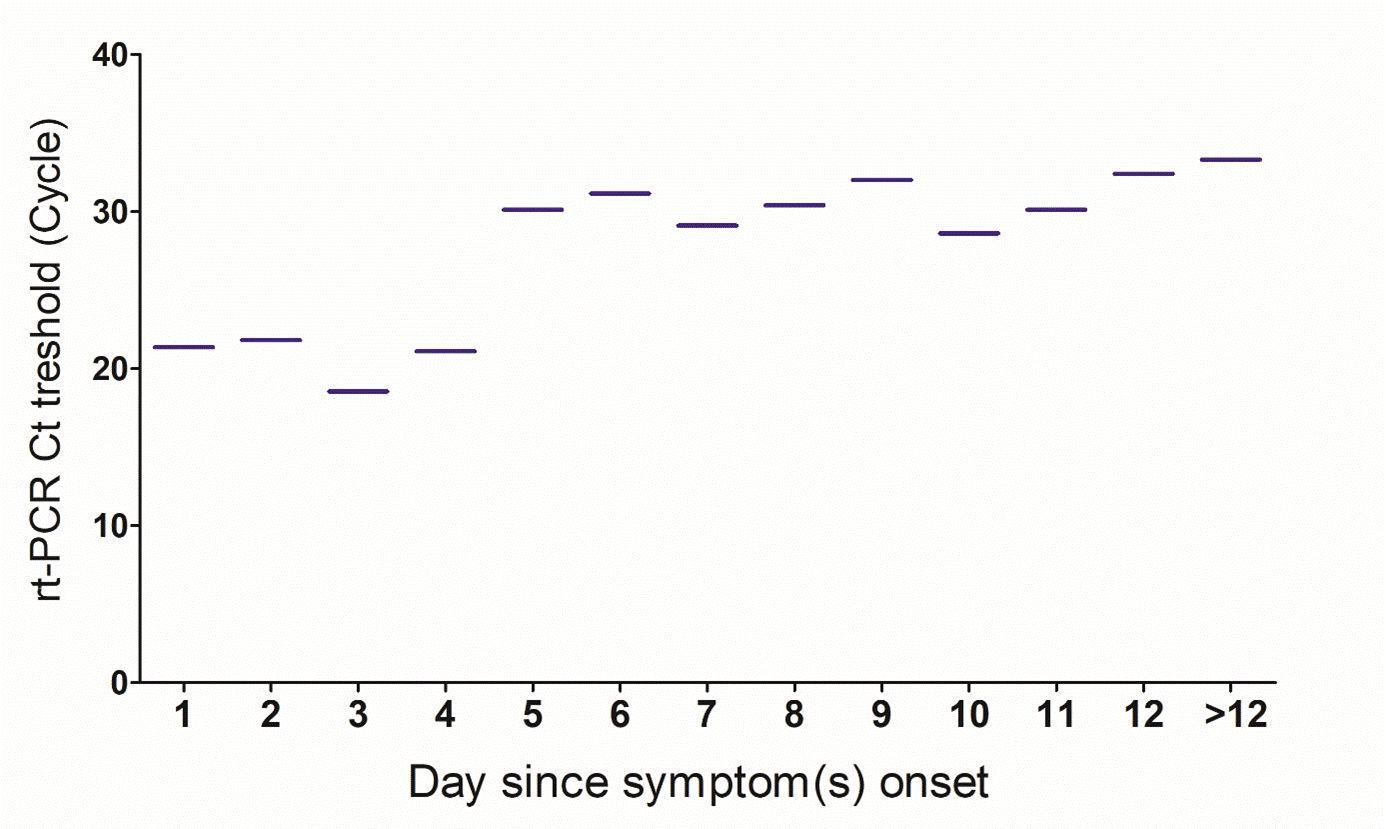
Figure 1: Level of the RT-PCR threshold cycle according to the day the symptom onset. Median of the RT-PCR threshold for days the symptom onset. Data are shown as median. Data has been analyzed using SPSS version 20 and graphing using GraphPad Prism 5.0.
Table 2. Clinical performance of the LumiraDx. Total study population size, the proportion of positive and negative RT-PCR and LumiraDx antigen SARS-CoV-2 results are displayed. The sensitivity, specificity, positive predictive value, negative predictive value and Cohen's kappa of the LumiraDx antigen test compared to RT-PCR are shown for the total study population and ranges of the RT-PCR threshold cycles. Statistical analysis was performed using SPSS version 20 (IBM, Inc.).
| |
LumiraDx Ag SARS-CoV-2 Test |
RT-PCR |
| |
All |
Ct ≤ 33 |
Ct>33 |
|
N |
184 |
175 |
150 |
184 |
Positive, % |
36 (19.7) |
33 (18.5) |
3 (2.0) |
44 (23.9) |
Negative, % |
150 (81.5) |
142 (81.5) |
147 (98.0) |
140 (76.1) |
Sensitivity (95% CI) |
81.8 (66.8-91.3) |
94.4 (79.9-99.0) |
27.2 (7.3-60.6) |
NA |
Specificity (95% CI) |
100 (96.4-100) |
100 (96.6-100) |
100 (96.6-100) |
NA |
PPV (95% CI) |
100 (87.9-100) |
100 (87.3-100) |
100 (31.0-99.4) |
NA |
PNV (95% CI) |
94.5 (89.2-100) |
98.5 (94.4-99.7) |
94.5 (89.2-97.4) |
NA |
Cohen's kappa |
0.86 |
0.96 |
0.41 |
NA |
PPV: Positive Predictive Value; NPV: Negative Predictive Value; Ct: Threshold Cycle; CI: Confident Interval |
Rapid diagnostic testing for SARS-CoV-2 is crucial to limit the spread of the virus and manage infected patients. In the light of the urgent need to increase the SARS-CoV-2 testing especially in Africa where the COVID-19 pandemic is progressing, accurate antigen point-of-care testing should be the key to expand SARS-CoV-2 diagnosis. Several SARS-CoV-2 antigen tests have been developed to palliate to the challenges of RT-PCR testing et can provide results within minutes [9-12,14,15,17]. Although those are easy-to-use and affordable diagnosis tools, there are limited data on their performance.
From nasal swap samples, RT-PCR showed an estimated prevalence of 23.9% while LumiraDx SARS-CoV-2 antigen test elicited a prevalence 19.7%. This evaluation was conducted at hospital setting in patients presenting SARS-CoV-2-related symptoms, which may explain the high prevalence observed. When compared to RT-PCR, the LumiraDx SARS-CoV-2 antigen assay displayed an overall sensitivity of 81.8%. The Lumira company has reported sensitivity of 97.6% and specificity of 96.6% [24], eliciting higher sensitivity compared to our results although we found higher specificity. However, this should not migrate our data which is in line with studies showing that sensitivities reported by the manufactures are often low that on site evaluations [9,12-14,16-18]. Moreover, the company performance has been evaluated in patients whose period of symptoms occurred within 12 days since onset while period for which symptoms onset in our cohort was within 40 days. The performance found in our study meets the WHO performance requirements for the use of SARS-CoV-2 Ag-RDTs of ≥ 80% sensitivity and ≥ 97% specificity when compared to RT-PCR a reference assay [25].
Our study shows that the sensitivity of the LumiraDx SARS-CoV-2 antigen assay is dependent of the RT-PCR threshold cycle. Indeed, for CT below or equal 33 cycles, the sensitivity was 94.4% versus 27.2% for Ct values above 33 cycles. This is in line with the study of Drain et al. that found higher sensitivity for Ct values below 33 cycles [20]. Whether the low sensitivity for Ct value above 33 cycles is a limitation of the LumiraDx SARS-CoV-2 antigen test need to be determined before its implementation. We found that within 12 days of the onset of symptoms, the LumiraDx test elicited an agreement with RT-PCR from 88.3% to 100% and 85.7% when symptoms appear after 12 days. This may support that lower Ct values are likely detected after 12 days the symptom onset. When stratifying the RT-PCR threshold cycle of positive samples for days the symptom onset, there was a trend of higher median Ct from day 12, although the difference was not significant. Some authors have correlated the infectivity to high viral load (low RT-PCR Ct value) or viability of the virus in culture [26,27]. Successful isolation of the virus in culture has also shown to be correlated with Ct values below 33 cycles [28] or to decrease from day 10 days since symptom onset [29]. These altogether may suggest that the cut-off of 33 below which the LumiraDx SARS-CoV-2 antigen test is most performant likely represents non-viable virus in culture.
We here demonstrated that compared to RT-PCT, the LumiraDx antigen SARS-CoV-2 test displayed sensitivity that meets the WHO performance requirements for the use of SARS-CoV-2 antigen testing. Its sensitivity was even highly improved for RT-PCR Ct values up to Ct 33 cycles. Good agreement with RT-PCR was also found withing 12 days the symptom onset. This suggests that the LumiraDx SARS-CoV-2 platform might be suitable for identification and management of SARS-CoV-2 infection in clinic settings.
We gratefully thank the individuals participating in this study and the staff of IRESSEF involved in the SARS-CoV-2 testing.
This work was supported by the “West African Task Force for the Control of Emerging and Re-emerging Infectious Diseases” (WATER), and “Innovation in Laboratory Engineered Accelerated Diagnosis” (iLEAD) (Grant Nr. OPP1214434/INV-009631). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The views expressed are those of the authors, and the funders are not responsible for any use that may be made of the information contained herein.
MM, SM, AD conceived and designed the study. PABD, NKB, NL and GL performed the experiments. MM, PABD, NKB, GL, and NL recruited study participants and collected data. MM analyzed and interpreted the data. SM, AD, MM, EMM, DW, NL, GL contributed to reagents/materials/analysis tools. MM, ID, MG, MC, MD, AD, GL, NL participated to study design. MM, SB, participated to study coordination. MM, GL wrote/drafted the manuscript. YAD, DW, NL, GL, JKB, AP, OD, AD, AA, MN, DGN, PAD, AM, BM, MDSN, TND, EMM reviewed the manuscript critically for important intellectual content. SM, and AD approved the final version to be published. All authors approved the final version of the manuscript.
- World Health Organization (2020) Health emergencies, coronavirus disease (COVID-19) outbreak. [Accessed 21 October 2020].
- Zhu N, Zhang D, Wang W, Li X, Yang B, et al. (2020) China Novel Coronavirus Investigating and Research Team. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med 382: 727-733. [Crossref]
- Wu F, Zhao S, Yu B, Chen YM, Wang W, et al. (2020) Author Correction: A new coronavirus associated with human respiratory disease in China. Nature 580: E7. [Crossref]
- Mathuria JP, Yadav R, Rajkumar (2020) Laboratory diagnosis of SARS-CoV-2 - A review of current methods. J Infect Public Health 13: 901-905. [Crossref]
- WHO. Interin Guidance (2020) Laboratory testing for coronavirus disease (COVID-19) in suspected human cases. CDC C-NC-nR-TR-PDP CDC. 2020. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel.
- Emery SL, Erdman DD, Bowen MD, Newton BR, Winchell JM, et al. (2004) Real-time reverse transcription-polymerase chain reaction assay for SARS-associated coronavirus. Emerg Infect Dis 10: 311-316. [Crossref]
- Udugama B, Kadhiresan P, Kozlowski HN, Malekjahani A, Osborne M, et al. (2020) Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano 14: 3822-3835. [Crossref]
- Sheridan C (2020) Fast, portable tests come online to curb coronavirus pandemic. Nat Biotechnol 38: 515-518. [Crossref]
- Porte L, Legarraga P, Vollrath V, Aguilera X, Munita JM, et al. (2020) Evaluation of a novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int J Infect Dis 99: 328-333. [Crossref]
- Nagura-Ikeda M, Imai K, Tabata S, Miyoshi K, Murahara N, et al. (2020) Clinical Evaluation of Self-Collected Saliva by Quantitative Reverse Transcription-PCR (RT-qPCR), Direct RT-qPCR, Reverse Transcription-Loop-Mediated Isothermal Amplification, and a Rapid Antigen Test To Diagnose COVID-19. J Clin Microbiol 58: e01438-20. [Crossref]
- Krogh J (1989) Empyema of the gallbladder: a case with unusual presentation. Acta Chir Belg 89: 204-205. [Crossref]
- Linares M, Pérez-Tanoira R, Carrero A, Romanyk J, Pérez-García F, et al. (2020) Panbio antigen rapid test is reliable to diagnose SARS-CoV-2 infection in the first 7 days after the onset of symptoms. J Clin Virol 133: 104659. [Crossref]
- Scohy A, Anantharajah A, Bodéus M, Kabamba-Mukadi B, Verroken A, et al. (2020) Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J Clin Virol 129: 104455. [Crossref]
- Mertens P, De Vos N, Martiny D, Jassoy C, Mirazimi A, et al. (2020) LHUB-ULB SARS-CoV-2 Working Diagnostic Group. Development and Potential Usefulness of the COVID-19 Ag Respi-Strip Diagnostic Assay in a Pandemic Context. Front Med (Lausanne) 7: 225. [Crossref]
- Kohmer N, Toptan T, Pallas C, Karaca O, Pfeiffer A, et al. (2021) The Comparative Clinical Performance of Four SARS-CoV-2 Rapid Antigen Tests and Their Correlation to Infectivity In Vitro. J Clin Med 10: 328. [Crossref]
- Toptan T, Eckermann L, Pfeiffer AE, Hoehl S, Ciesek S, et al. (2021) Evaluation of a SARS-CoV-2 rapid antigen test: Potential to help reduce community spread? J Clin Virol 135: 104713. [Crossref]
- Mak GCK, Lau SSY, Wong KKY, Chow NLS, Lau CS, et al. (2021) Evaluation of rapid antigen detection kit from the WHO Emergency Use List for detecting SARS-CoV-2. J Clin Virol 134: 104712. [Crossref]
- Nalumansi A, Lutalo T, Kayiwa J, Watera C, Balinandi S, et al. (2021) Field evaluation of the performance of a SARS-CoV-2 antigen rapid diagnostic test in Uganda using nasopharyngeal samples. Int J Infect Dis 104: 282-286. [Crossref]
- World Health Organization (2020) A coordinated global research roadmap: 2019 novel coronavirus. Available at: https://www.who.int/blueprint/priority-diseases/key-action/Coronavirus_Roadmap_V9.pdf. [Accessed 21 October 2020].
- Drain PK, Ampajwala M, Chappel C, Gvozden AB, Hoppers M, et al. (2021) A Rapid, High-Sensitivity SARS-CoV-2 Nucleocapsid Immunoassay to Aid Diagnosis of Acute COVID-19 at the Point of Care: A Clinical Performance Study. Infect Dis Ther 10: 753-761. [Crossref]
- LumiraDx (2020) LumiraDx website and SARS-CoV-2 Antigen test EUA Product Insert. Available at: https://www.lumiradx.com/us-en/ [Accessed 21 October 2020].
- Lai MM (2003) SARS virus: the beginning of the unraveling of a new coronavirus. J Biomed Sci 10: 664-675. [Crossref]
- Instructions for Use. Available at: https://www.fda.gov/media/137178/download. 2020
- LumiraDx. Performance evaluation of the LumiraDx SARS-CoV-2 Antigen Test to aid diagnosis of acute COVID-19 at the point of care. Available at: https://www.lumiradx.com/assets/pdfs/white-papers/performance-evaluation-of-sars-cov-2-ag-test.pdf?v=1. S-COM-ART-00865 R1 Date of Revision 2020/11
- WHO Antigen-Detection in the Diagnosis of SARS-CoV-2 Infection Using Rapid Immunoassays: Interim Guidance. Available at: https://www.who.int/publications/i/item/antigen-detection-in-the-diagnosis-of-sars-cov-2infection-using-rapid-immunoassays [Accessed 21 December 2020].
- Perera RAPM, Tso E, Tsang OTY, Tsang DNC, Fung K, et al. (2020) SARS-CoV-2 Virus Culture and Subgenomic RNA for Respiratory Specimens from Patients with Mild Coronavirus Disease. Emerg Infect Dis 26: 2701-2704. [Crossref]
- van Kampen JJA, van de Vijver DAMC, Fraaij PLA, Haagmans BL, Lamers MM, et al. (2021) Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19). Nat Commun 12: 267. [Crossref]
- La Scola B, Le Bideau M, Andreani J, Hoang VT, Grimaldier C, et al. (2020) Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis 39: 1059-1061. [Crossref]
- Wolfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, et al. (2020) Virological assessment of hospitalized patients with COVID-2019. Nature 581: 465-469. [Crossref]
Editorial Information
Editor-in-Chief
Yeun-Hwa Gu
Junshin Gakuen University, Japan
Article type
Research Article
Publication History
Received: July 05, 2021
Accepted: August 20, 2021
Published: August 27, 2021
Copyright
©2021 Mbow M. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation
Mbow M, Dieye PAB, Ba NK, Cisse M, Lo G, et al. (2021) Evaluation an immunofluorescence-based antigen test for hospital point-of-care diagnosis of SARS-CoV-2 infection. Clin Microbiol Infect Dis 6: DOI: 10.15761/CMID.1000192.

