Abstract
This article presents a molecular approach to measure the magnitude of the expression of 88 key micro(mi)RNA genes isolated from the stool of healthy human adults following intake of Pomegranate juice (PGJ), functional fermented sobya (FS), rich in potential probiotic lactobacilli, or their combination. Total small RNA was isolated from stool of 25 volunteers before and following a three-week dietary intervention. Expression of 88 miRNA genes was evaluated using 96 well plate RT2 miRNA qPCR arrays. Using parallel coordinates plots, there was hardly any separation for the gene expression (Cq) values, and none of the miRNAs showed significant statistical expression after controlling the false discovery rate. Melting temperature profiles produced during PCR amplification, however, found seven significant genes (miR-184, miR-203, miR-373, miR-124, miR-96, miR-373 and miR-301a), which separate candidate miRNAs that could serve as novel biomarkers of relevance to oxidative stress and immunoglobulin function for the intake of polyphenol (PP)-rich, functional fermented foods rich in lactobacilli (FS), or their combination.
Key words
biomarkers, fermented sobya, pomegranate, PCR
Introduction
Disease modulation by nutrients
The WHO in 2011 predicted that by 2030, cardiovascular diseases will remain the leading causes of death, affecting approximately 23.6 million people worldwide [1]. Hypercholesterolemia represents the first stage of atherosclerosis, a risk factor for Chronic Heart Disease (CHD) [2]. The prevalence of chronic, degenerative diseases attributable wholly or partially to dietary patterns is the most serious threat to public health [3]. In addition, research suggests that about one-third of all cancer deaths are attributable to poor nutrition, physical inactivity, and overweight or obesity; and these risk factors may account for up to 80% of large bowel, breast, and prostate cancers. A common factor that may contribute to the development and progression of these illnesses is chronic inflammation [4], which can be caused and modified by diet.
Pomegranate juice (PGJ) and derived products are amongst the richest natural sources of polyphenolic compounds, with positive implication on plasma lipid profile, i.e., TC, LDL-C and TG [5]. Furthermore, the presence of anthocyanin & ellagitannins pigments, mainly punicalagins, inhibits the activities of 3-hydroxy-3-methylglutaryl-CoA reductase and sterol O-acyltransferase, two key enzymes in cholesterol metabolism [6]. Probiotic bacteria also contribute to lowering plasma hyper cholestrolemia due to the proposed mechanism mediated by the probiotic bile salt hydrolase (BSH) activity. This probiotic enzyme hydrolyses conjugates glycodeoxycholic acid and taurodeoxycholic acid to hydrolysis products, which inhibit the absorption of cholesterol and suppress the reabsorption of bile acid [7].
The colonic microbiota is a central site for the metabolism of dietary PP and colonization of probiotic bacteria. A dietary intervention study with probiotic strains from the species Lactobacillus acidophilus, L. casei and L. rhamnosus given to healthy adults showed that the consumption of bacteria resulted in the differential expression of from several hundred up to thousands of genes in vivo in human mucosa, elucidating how intestinal mucosa of healthy humans perceives different probiotics and provides avenues for rationally designed tests of clinical applications [8]. The interaction of PP with the gut microbiota influences the expression of selected human genes (i.e., nutritional transcriptomics), which mediates mechanisms underlying the beneficial health effects [9]. Similar in vivo mucosal transcriptome findings have also been reported when adults were given the probiotic Lactobacilus plantarum, illustratinging how probiotics may modulate human cellular pathways and uncovered remarkable similarity to response profiles obtained for specific bioactive molecules and drugs [8].
Gene expression and its control by miRNAS
The gene expression profile of a cell determines its function, phenotype and cells' response to external stimuli, and therefore help elucidate cellular functions, biochemical pathways and regulatory mechanisms [10]. During the past years, several gene expression profiling methods at the RNA level have emerged and applied successfully to cancer research. Profiling by microarrays allows the parallel quantification of thousands of genes from multiple samples simultaneously using a single RNA preparation and has become important because microarrays are convenient to use, and do not require large-scale DNA sequencing, provides a clear picture of cells' physiological state, and appears to be a comprehensive approach to characterize cancer molecularly, as seen in studies on colon cancer [10].
Effect of antioxidant polyphenols abundant in Mediterranean diets on gene expression unraveled by the availability of molecular biology techniques, reveals our adaptation to environmental changes [11]. Efforts to characterize the human transcriptome have largely been applied to tissue, blood, and urine (i.e., normally sterile materials), as well as stool (a non-sterile medium). Extraction protocols using commercially available reagents to obtain high-yield, reverse-transcribable RNA from human stool in studies performed on colon cancer have been reported [10,12].
Micro(mi)RNAs as biomarkers, and their roles in disease processes
A biomarker is a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacological responses to a therapeutic intervention. In contrast, clinical endpoints are variables that represent a study subject's health from the subject's perspective. A variety of biomarkers exist today as surrogates to access clinical outcomes in diseases, predict the health of individuals, or improve drug development. An ideal biomarker should be safe and easy to measure, cost effective to follow up, modifiable with treatment, and consistent across gender and ethnic groups. Since we never have a complete understanding of all processes affecting individual's health, biomarkers as surrogate endpoints need a constant reevaluation of the relationship between surrogate endpoints and a true clinical endpoint [13]. In the present study, we have used microRNAs as biomarkers for the intake of PP-rich or fermented foods.
Gene expression and its control by miRNAs
The gene expression profile of a cell determines its function, phenotype and cells' response to external stimuli. Therefore, gene expression profiles can help elucidate cellular functions, biochemical pathways and regulatory mechanisms. During the past few years, several gene expression profiling methods at the RNA level have emerged and have been applied successfully to cancer research [10,12].
Control of gene expression has also been studied by miRNA molecules, a small non-coding RNA molecule (18–24 nt long), involved in transcriptional and post-transcriptional regulation of gene expression by inhibiting gene translation. MiRNAs silence gene expression through inhibiting mRNA translation to protein, or by promoting the degradation of mRNA. Since first reported in the literature in 1993 [14], the number of identified miRNAs in June 2014, version 14.0, the latest miRBase release (v20) [15] contains 24,521 miRNA loci from 206 species, processed to produce 30,424 mature miRNA products. MiRNAs are processed by RNA polymerase II to form a precursor step which is a long primary transcript. Pri-miR is converted to miRNA by sequential cutting with two enzymes belonging to a class of RNA III endonucleases, Drosha and Dicer enzymes. Drosha converts the long primary transcripts to about ~70 nt long primary miRNAs (pri-miR), which are then exported to the cytoplasm by Exportin 5, converted to mature miRNA (~22 net) by Dicer [16]. Each miRNA may control multiple genes, and one or more miRNAs regulate a large proportion of human protein-coding genes, whereas each single gene may be regulated by multiple miRNAs [17]. MiRNAs inhibit gene expression through interaction with 3-untranslated regions (3 UTRs) of target mRNAs carrying complementary sequences [18].
Materials and methods
Participants
The study subjects were 25 healthy adults, with age ranging between 20 – 34 years, without metabolic diseases, not using any medication for the last 6 weeks, and with no signs of allergy or hypersensitivity to any food or ingested material. Compliance with the supplementation in all subjects was satisfactory as assessed by daily contacts. All subjects continued their habitual diets during the study. The research protocol received approval from the institution review board and all subjects provided written, informed consent before participation in the study.
Design of the study
Figure 1 represents the design of the randomized controlled trial (RCT). Estimated dietary intake was assessed by 3 repeated food records, one week before enrollment in the trial. The average portion size consumed and composition data values from nutrient composition of the food were combined to assess average daily energy and nutrient intakes by the “nutrisurvey” software program. The characteristics of the voluntary subjects who were enrolled in the study, the mean daily energy intake, as well as selected macro nutrients are presented in Table 1.
Table 1. Composition of the supplements
Parameter |
Unit |
Dietary supplements |
Control
(Portion served) |
FS
(portion served) |
PGJ
(portion served) |
FS+PGJ
(portion served) |
Portion Size |
g |
– |
170 |
250 |
150 + 10 |
Total Solids |
g |
– |
40.01 |
17.75 |
48 |
Carbohydrate |
g |
– |
51.1 |
32.75 |
59 |
Dietary Fiber |
g |
– |
54 |
0.25 |
48.6 |
Energy |
kcal |
– |
263 |
135 |
290 |
Lactobacillus |
diverse cfu/serving size |
– |
– |
5.1×109 |
4.5×109 |
Yeast |
cfu/serving size |
– |
– |
2.77×1010 |
2.44×1010 |
Total PP |
mg*/portion g |
– |
– |
519.1±8.75 |
207.65±3.5 |
Antioxidantactivity |
(AEAC)** |
– |
7.74±1.33 |
11.35±2.2 |
11.37±2.2 |
*mg galic acid equivalent (GAE), FS: Functional fermented sobya; PGJ: Pomegranate juice
**mmol ascorbic acid equivalent antioxidant activity.
Modified from Reference 5 Gouda et al. [5].
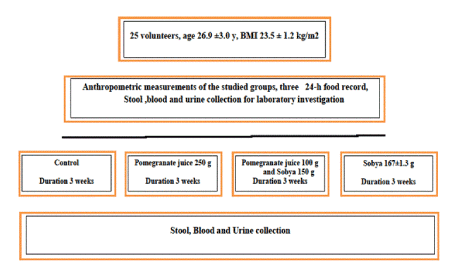
Figure 1. A chart illustrating the design of randomized control study
Supplements
Pomegranate was purchased in bulk from the Obour Public Market, Cairo, Egypt. Pomegranate fruits were peeled and the juice was obtained using a laboratory pilot press (Braun, Germany). The juice was distributed in aliquots of 100 or 250 grams in air tight, light-proof polyethylene bottles, and saved at −20°C. Under this storage condition, the pomegranate polyphenols were fully stable. Sour sobya, a fermented porridge was purchased twice a week from the retail market and saved in the refrigerator. Sobya is fermented rice containing per gram 3 × 107 cfu diverse lactic acid bacteria (LAB) and 1×107 cfu Sacharomyces cerivisiae. Other ingredients: added milk, sugar and grated coconut. Table 2 presents the proximate initial and final mean urinary polyphenols, plasma and urinary antioxidative activity, urinary thiobarbituric acid reactive species (TBARS), and erythrocytic glutathione-S-transferase (GST).
Table 2. Initial and final mean urinary polyphenols, plasma and urinary antioxidative activity, urinary TBARS and erythrocytic GST
Parameter |
Unit |
Control |
Sobya |
|
Pomegranate |
Sobya+Pomegranate |
| |
|
Baseline |
Final |
Baseline |
Final |
|
Baseline |
Final |
Baseline |
Final |
| |
|
X±SE |
X±SE |
X±SE |
X±SE |
p |
X±SE |
X±SE |
p |
X±SE |
X±SE p |
Urinary polyphenol |
GAE/mg |
10.36 |
8.11 |
11.84 |
9.86 |
|
5.70 |
55.23 |
<0.05 |
10.40 |
21.62 |
creat |
±1.8 |
±2.2 |
±6.2 |
±1.8 |
|
±1.4 |
±21.7 |
|
±3.2 |
±7.3 |
Urinary antioxidant activity |
AEAC/mg |
9.74 |
8.13 |
3.89 |
10.30 |
|
7.18 |
46.57 |
<0.05 |
10.90 |
20.25 |
creat* |
±2.0 |
±2.7 |
±09 |
±2.3 |
|
±0.9 |
±18.0 |
|
±2.4 |
±3.9 |
Urinary TBARS |
ug/mg |
83.04 |
75.17 |
82.77 |
29.97 |
|
173.93 |
51.48 |
<0.05 |
157.70 |
40.62 |
creat |
±12.1 |
±15.3 |
±27.8 |
±4.4 |
|
±44.8 |
±8.2 |
|
±47.8 |
±8.3 |
Plasma antioxidant |
AEAC/ |
6.36 |
5.99 |
3.70 |
4.55 |
|
3.64 |
5.92 |
<0.05 |
2.78 |
4.49 |
1oo ml |
±2.81 |
±2.66 |
±0.33 |
±0.27 |
|
±0.30 |
±0.68 |
|
±0.11 |
±0.58 |
E-GST activity |
IU/g Hb |
5.94 |
5.45 |
4.26 |
7.21 |
<0.05 |
4.73 |
8.34 |
|
4.56 |
6.90 |
| |
±3.3 |
±4.1 |
±0.5 |
±0.8 |
|
±1.0 |
±1.0 |
|
±1.0 |
±1.0 |
X±SE: Mean ± Standard Error, *mmol ascorbic acid equivalent antioxidant capacity/mg creatinine; GAE: gallic acid equivalent, Mean values are significantly different if the P-values are less than 0,05 (P<0.05). Modified from Reference 5 Gouda et al. [5]
Stool collection and storage
Stool was obtained from the 25 healthy adults, twice at day 0 and three weeks after the dietary intervention. All stools were collected with a sterile, disposable wood spatulas from clean containers, after the stools were freshly passed, and then placed for storage into Nalgene screw top vials (Thermo Fisher Scientific, Inc., Palo Alto, CA, USA), each containing 2 ml of the preservative RNA later (Applied Biosystems/Ambion, Austin, TX, USA), which prevents the fragmentation of the fragile mRNA [10], and vials were stored at – 70°C until analysis. Total small RNA, containing miRNAs, was extracted from all frozen samples at once, when ready, and there was no need to separate mRNA containing small miRNAs from total RNA, as small total RNA was suitable to make ss miRNA c-DNA.
Extraction of total small RNA
A procedure that we used for extracting small total RNA from stool was carried out using a guanidinium-based buffer, which comes with the RNeasy isolation Kit®, Qiagen, Valencia, CA, USA, as we have previously detailed [10] DNase digestion was not performed, as our early work has demonstrated no difference in RNA yield or effect on RT-PCR after DNase digestion. The time to simultaneously purify aqueous RNA from all of the 25 frozen stool samples from frozen samples was ~three hours. Small RNA concentrations were measured spectrophotometrically at λ 260 nm, 280 nm and 230 nm, using a Nano-Drop spectrophotometer (Themo-Fischer Scientific). The integrity of total RNA was determined by an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Palo Alto, CA, USA) utilizing the RNA 6000 Nano LabChip®. RNA integrity number (RIN) was computed for each sample using the supplied software.
Preparation of ss-cDNA for molecular analysis
The RT2 miRNA First Strand Kit® from SABiosciences Corporation (Frederick, MD, USA) was then employed for making a copy of ss-DNA in a 10.0 µl reverse transcription (RT) reaction, for each RNA samples in a sterile PCR tube, containing 100 ng total RNA, 1.0 µl miRNA RT primer & ERC mix, 2.0 /µl 5X miRNA RT buffer, 1.0 µl miRNA RT enzyme mix, 1.0 µl nucleotide mix and Rnase-free H2O to a final volume of 10.0 µl. The same amount of total RNA was used for each sample. Contents were gently mixed with a pipettor, followed by brief centrifugation. All tubes were then incubated for 37° for 2 hours, followed by heating at 95° for 5 minutes to degrade the RNA and inactivate the reverse transcriptase. All tubes were chilled on ice for 5 minutes, and 90 µl of Rnase-free H2O was added to each tube. Finished miRNA First Strand cDNA synthesis reactions were then stored overnight at -20°.
Using a cancer RT2 miRNA PCR array 96-well plate to study miRNAs' expressions
We used a SABiosciences RT2 miRNA qPCR Array Plate System for Human (Qiagen) to analyze miRNA expression using real-time, reverse transcription PCR (RT-qPCR) as a very sensitive and reliable quantitative method for miRNA expression analysis. The arrays take advantage of a SYBR Green real-time PCR detection system, which has been optimized to analyze the expression of many mature miRNAs simultaneously. Each 96-well array plate contains a panel of primer sets for 88 relevant miRNA focused pathways (one universal primer and one gene-specific primer for each miRNA sequence), plus four housekeeping genes (Human SNORD 48, 47 and 44, and U6), and two RNA and two PCR quality controls. Duplicate RT Controls (RTC) to test the efficiency of the miRNA RT reaction, with a primer set that detects the template synthesized from the built in miRNA External RNA Control (ERC). There are duplicate RT controls (RTC) to test the efficiency of the miRNA reverse transcription process, with a primer set to detect the template synthesized from the kit's built-in miRNA External RNA Control (ERC). There is also duplicate positive PCR controls (PPC) to test the efficiency of the PCR process, using a per-dispensed artificial DNA sequence and the primer set that detects it. The two sets of duplicate control wells (RTC and PPC) also test for inter-well, intra-plate consistency. The human RT2 miRNA PCR Arrays reflect miRNA sequences annotated by the Sanger miRBase Release 14. Refer Figure 2 for the layout of the MAH-102F array.

Figure 2. A diagram illustrating the design of the randomized controlled trial
Performing real-time quantitative polymetrase chain reaction (qPCR)
We used RT2 SYBR Green qPCR Master Mix (SBA Biosciences) for obtaining accurate results from our PCR arrays. We mixed the following components in a 15-ml tube for 96-well plate format: 1275 µl of 2X RT2 SYBR Green PCR Master Mix, 100 µl of diluted first strand reaction, 1175 µl of ddH2O (total volume 2550 µl, of which 2400 was needed for 96 reactions, each well having 25 µl, with150 µl cocktail remaining.
We employed a Roche LightCycler 480® 96-well block PCR Machine (Roche, Mannheim, Germany). When ready, we removed the needed miRNA qPCR Arrays, each wrapped in aluminum foil, from their sealed bags, added 25 µl of the same cocktail to each well, adjusted the ramp rate to 1°C/sec. We used 45 cycles in the program to allow the use of the Second Derivative Maximum method, available with the LightCycler 480® software for data analysis [19]. We first heated the 96 well plate for 10 min at 95°C to activate the HotStart DNA polymerase, then we used a three-step cycling program (a 15 seconds heating at 95° to separate the double stranded DNA, a 30 seconds annealing step at 60° to detect and record SYBR Green fluorescence at each well during each cycle, and a final heating step for 30 seconds at 72°). Each plate was visually inspected after the run for signs of evaporation from the wells. Data were analyzed using the 2-ΔΔCt method [20]. Resulting threshold cycle values for all wells were exported to a blank Excel sheet for data analysis. We also
ran a Dissociation (Melt) Curve Program after the cycling program [21] and generated a first derivative dissociation curve for each well in the plate, using the Lightcycler's® software.
Statistical analysis
Gene expressions were standardized by dividing by the SNORD48 value while raw melting temperatures were used. Analysis were done using the software R (version 3.1.3), with the package MASS [22]. For each standardized gene and each melting temperature a one way ANOVA was used to obtain a p-value. There were four levels of the explanatory variable: Control, Sobya, Pom, and Both. Parallel coordinate plots (parcoord command in R) [23] were used to visualize the data for each gene and each melting temperature. Coordinates were ordered using the magnitude of the p-value. The two sample t-test was used on gene expression to compare Control to Sobya and Control to Both (t.test command in R with var.equal=FALSE). P-values were adjusted to control for false discovery rate. The method is described in [24] Benjamini and Yekutieli (p.adjust command in R with method=‘BY’).
Results
At base line, all participants in the trial excreted urinary total polyphenols; however, the inter individual variation was considerably high (4.89-12.59 mg GAE/100 ml urine).
Composition of the three supplements (FS, PGJ and FS + PGJ served to the volunteers is presented in Table 1. The initial and final mean urinary polyphenols, plasma and urinary antioxidative activity, urinary TBARS and erythrocytic GST. The daily portion of PGJ provided 21 mg PP /day, and the combination of PGJ – FS was 9 mg PP /day, is presented in Table 2.
Figure 3 is a lay out of RT2 miRNA PCR Array Human Cancer microRNA (MAH-102A). Figure 4 is a graphical representation of the parallel plot coordinates of the studied miRNA genes for melting temperature curve analysis. The genes were ordered using the p-values of a one way ANOVA based on groups. Genes with the smallest p-values are presented first.

Figure 3. Array lay out for human cancer RT2 miRNA qPCR Plate Array (MAH-102A)
Figure 4a show control genes (Snord48, Snord47, Snord44, RNUU6-2. MiRTC1, miRTC2, and PPC1, PPC2). In Figure 4b, five miRNA genes show separation. Gene miR-184 has the highest separation from the control gene. MiR-203 genes is hardly amplified in Sobya, while it is highly expressed in Pomegranate. For miR-373 gene, the control group is different from the other three treatment groups. For genes miR-124, miR-96 and miR-378, Pomegranate is well separated from other three groups. In Figure 4c, for gene miR-301a, the control is separated from the other three groups. Additional miRNA genes are not shown, as their p-values are greater (less significant), and the graphs did not show any meaningful separations.
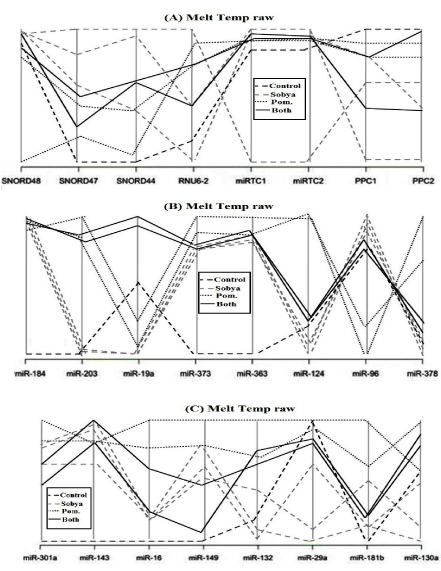
Figure 4. A graphical representation of the parallel plot coordinates of the studies miRNA genes. The genes were ordered using the p-values of a one-way ANOVA based on groups. Genes with the smallest p-values are presented first. Figure 4a show control genes. In Figure 4b, five miRNA genes show separation
Discussion
Suitability of stool as a medium for developing a sensitive molecular biomarker screen
Stool represents a challenging environment, as it contains many substances that may not be consistently removed in PCR, in addition to the presence of certain inhibitors, which all need to be removed for a successful PCR reaction. Our results [10,25] and others [12,26] have shown that the presence of bacterial DNA, non-transformed RNA and other substances in stool do not interfere with measuring miRNA expressions, especially when suitable PCR primers are employed for quantitative measurement. Besides, stool colonocytes contain more miRNA and mRNA than that available in free circulation, as in plasma [27], facilitating measurements.
PCR amplification and the effect of inhibitory substances
PCR is used for characterization of genetic markers because of its extreme sensitivity. This method, however, could lead to errors because of the presence of inhibiting substances, representing diverse compounds with different properties and mechanisms of action, which exert their effects through direct interaction with DNA that will be amplified, or through interference with the employed thermostable DNA polymerase [28]. Magnesium is a critical cofactor, and agents that reduce Mg2+ availability, or interfere with the binding of Mg2+ to DNA polymerase could inhibit the PCR reaction [29]. Calcium ion is another inorganic inhibiting substance, although most PCR inhibitors are organic compounds (e.g., bile salts, sodium dodecyl sulphate, urea, phenol, ethanol, polysaccharides), as well as proteins (e.g., collagen, hemoglobin, immunoglobin G and protineases) [30]. The existence of polysaccharides in stool could reduce the capacity to resuspend precipitated RNA or disrupt the enzymatic reaction by mimicking the structure of nucleic acid. The DNA template of the PCR can be inhibited by nucleases, and primers binding to DNA template my be interfered with by inhibitors [31]. Remedial strategies for removal of inhibitors in stool have been suggested such as additional extraction steps, sephadex G-200 chromatography, heat treatment before the PCR, chloroform extraction, treatment with activated carbon, adding BSA, or dilution of the sample [32]. In our hands, we have found that the dilution method, in which the extracted ribonucleic acid (RNA) is diluted in the reaction mixture, to be the most practical method for preventing PCR inhibition using a commercially available diluent [33].
Role of miRNAs in nutritional diseases and therapy
MiRNA functions seem to regulate development [34] and apoptosis [35]. Just as miRNA is involved in the normal functioning of eukaryotic cells, so has dysregulation of miRNAs been associated with many diseases such as various cancers [36], heart diseases [37], kidney diseases [38]. nervous system diseases [39], alcoholism [40], obesity [41], auditory diseases [42], eye diseases [43], skeletal growth defects [44], as well as key role in host–virus pathogenesis of viral diseases [45]. A negative correlation was found between tissue specificity of interactions and miRNA in a number of diseases, and an association between miRNA conservation and disease, as well as the finding that miRNAs associated with the same disease trend to emerge as predefined miRNA group,will collectively allow for identification of novel disease biomarkers at the miRNA level [46]. Specific miRNAs are critical in oncogenesis [47], effective in classifying solid [48] and liquid tumors [49] and serve as oncogenes or tumor suppressor genes [50]. MiRNA genes are frequently located at fragile sites, as well as minimal regions of loss of heterozygosity, or amplification of common breakpoints regions, suggesting their involvement in carcinogenesis [51]. MiRNAs have promise to serve as biomarkers for cancer diagnosis, prognosis and/or response to therapy [52,53]. Profiles of miRNA expression differ between normal tissues and tumor types, and evidence suggests that miRNA expression profiles can cluster similar tumor types together more accurately than expression profiles of protein-coding messenger (m)RNA genes [54]. Besides, small miRNAs (~18-22 nt long) are stable molecules than the fragile mRNA [25].
Role of melting curve analysis in elucidating preferential miRNA expression
Melt curve analysis is an assessment of the dissociation characteristics of ds DNA during heating. The use of DNA-binding dyes, as SYBR Green I, which fluoresces 1000-fold more intensely while intercalated in the minor grooves of the two strands of DNA, the dissociation of the DNA during heating is measured by the large reduction in fluorescence that results, a melt curve profile can be generated, and specialized PCR systems as Roche LightCycler 480® used in this study records the total fluorescence generated by SYBR Green I binding to the ds DNA as temperature changes, and plots the fluorescence in real time, as a function of temperature [55].The first derivative of this plot (dF/dT) is the rate of change of fluorescence in the reaction, and a significant change in fluorescence accompanies the melting of the ds PCR product. A plot of -df/dT versus temperature will display these changes in fluorescence as distinct peaks, and makes it easier to pin-point the temperature of dissociation (defined as 50% dissociation) by virtue of the peaks formed. The melting temperature (Tm) of each product is defined as the temperature at which the corresponding peak maximum occurs. This analysis confirms the specificity of the chosen primers, as well as reveal the presence of primer dimers, which usually melt at lower temperatures than the desired product, because of their small size, and their presence severely reduce the amplification efficiency of the target gene since they compete for reaction component during amplification and ultimately the accuracy of the data. The greatest effect is observed at the lowest concentrations of DNA, which ultimately compromises the dynamic range. Moreover, nonspecific amplifications may result in PCR products that melt at temperatures above or below that of the desired product. Optimizing reaction components (Mg2+, detergents, SYBR Green I concentration) and annealing temperatures aid in decreasing nonspecific product formation, but proper product design is the best method to avoid nonspecific products. Including a negative control will determine if there is a coamplified genomic DNA [55,56]. High resolution melting (HRM) is an advancement of this general technology to identify variation in nucleic acid sequences enabled by a more advanced software, and is less expensive than probe-based genotyping methods, and allows identification of variants quickly and accurately [57]. This method has been widely used for detection of bacteria [58,59] viruses [60], mutations [61] and translocations [62]. In our study, we found the melt curve analysis to be a useful and an informative method, and we think that it is imperatives for investigators to run this kind of analysis on samples that do not show expression differences in their mRNA or miRNA studied genes.
The clinical significance of the data presented above is that using melting temperatures for analyzing nutrient-gene data is a promising new approach for identifying key regulatory miRNA genes related to metabolites rich in polyphenols, probiotic lactobacilli, or combinations of the two metabolites. Melt curve analysis is a powerful novel approach because after the statistical analysis carried on our miRNA samples produced negative results, running melt curve analysis on the same samples distinguished 7 of the 88 miRNA genes imprinted on the highly sensitive focused PCR arrays (~8% of the genes) to be significant. Thus, we believe that it is imperatives for investigators to run this kind of “melt curve” analysis on nutrition samples that are mild in nature, and many not show significant differences in the expression of studied miRNA genes. The same analysis can also be envisioned for messenger RNA amplifications, using mRNA arrays. We are also planning to carry out additional miRNA expression studies with much more observations using PP, FS and their combinations, and collectively the obtained results would fully demonstrate the sensitivity/specificity of this powerful systemic molecular approach for analyzing nutrient-gene data.
Acknowledgments
We express our deep gratitude to those volunteers who participated in this research by completing a dietary questionnaire, and providing urine, blood and stool samples for the study.
The Authors declare no conflict of interest in carrying out this research or preparing this article for publication.
References
- World Health Organization (WHO) (2011) Cardiovascular Diseases (CVDs), Who Fact Sheet No. 317.
- Werner RM, Pearson TA (1998) LDL-cholesterol: a risk factor for coronary artery disease--from epidemiology to clinical trials.Can J Cardiol14 Suppl B: 3B-10B. [Crossref]
- Bernedetti S, Catalani S, Palma F, Trari FC (2011) The antioxidant protection of CELLFOOD® against oxidatice change in vitro. Food Chem Toxicol 49: 2292-2298.
- Bengmark S (2007) Advanced glycation and lipoxidation end products--amplifiers of inflammation: the role of food. J Parenter Enteral Nutr 31: 430-40.
- Gouda M, Moustafa A, Hussein L, Hamza M (2016) Three-week dietary intervention using apricots, pomegranate juice or/and fermented sour sobya and impact on biomarkers of antioxidative activity, oxidative stress and erythrocytic glutathione transferase activity among adults. Nutritional J 15: 52.
- Esmaillzadeh A, Tahbaz F, Gaieni I, Alavi-Majd H, Azadbakht L (2006) Cholestrol-lowering effect of concentrated pomeg- ranate juice consumption in Type II diabetic patients with hyperlipidemia. Int J Vit Nutr Res 76: 147-151. [Crossref]
- Zhuang G, Liu XM, Zhang QX, Tian FW, Zhang H, et al. (2012) Research advances with regards to clinical outcome and potential mechanisms of the cholestrol-lowering effects of probiotics. Clin Lipidol 7: 501-507.
- van Baarlen P, Troost F, van der Meer C, Hooiveld G, Boekschoten M, et al. (2011) Human mucosal in vivo transcriptome responses to three lactobacilli indicate how probiotics may modulate human cellular pathways. Proc Natl Acad Sci USA 108: 4562-4569. [Crossref]
- Richards AL, Burns MB, Alazizi A, Barreiro LB, Pique-Regi R, et al. (2016) Genetic and transcriptional analysis of human host response to healthy gut microbiota.mSystems1. [Crossref]
- Ahmed FE, Vos P, IJames S, Lysle DT, Allison RR, et al. Transcriptomic molecular markers for screening human colon cancer in stool and tissue. Cancer Genom Proteom 4: 1-20. [Crossref]
- De Caterina R, Madonna R (2004) Nutrients and gene expression.World Rev Nutr Diet93: 99-133. [Crossref]
- Link A, Balageur F, Shen Y, Nagasaka T, Lozano JJ, et al. (2010) Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol Biomarkers Prev 9: 1766-1774. [Crossref]
- Strimbu K, Tavel JA (2010) What are biomarkers? Curr Opin HIV AIDS 5: 463-466.
- Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes a small RNAs with antisense complimentarity to lin-14. Cell 75: 843-854.
- Kozomara A, Griffithis JS (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68-D73.
- Lund E, Dahlberg JE (2006) Substrate selectivity of exportin 5 and Dicer in the biogenesis of microRNAs. Cold Spring Harb Symp Quant Biol 71: 59-66.
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, et al. (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–73. [Crossref]
- Biomarkers Definition Working Group (2001) Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Therapeutics 69: 89–95.
- Luu-The V, Paquet N, Calvo E, Cumps J (2005) Improved real-time RT-PCR measurements using second derivative calculation and double correction. Biotechniques 38: 287-293. [Crossref]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method.Methods25: 402-408. [Crossref]
- Ririe KM, Rasmussen RP, Wittwer CT (1997) Product determination by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem 245: 154-160.
- Venables WN, Ripley BD (Eds.) (2002) Modern and Applied Stastistics. Fourth Edition. Springer, New York.
- R Core Team (2015) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Australia. http://www.R-project.org/
- Benjamini Y, Yekutieli D (2001) The controil of the false discovery rate in multiple testing under dependency. Annals Stat 28: 1165-1188.
- Ahmed FE, Ahmed NC, Vos PW, Bonnerup C, Atkins JN, et al. (2013) Diagnostic microRNA markers to screen for sporadic human colon cancer in stool: I. Proof of principle.Cancer Genomics Proteomics10: 93-113. [Crossref]
- Davidson LA, Lupton JR, Miskovsky E, Fields AP, Chapkin RS (2003) Quantification of human intestinal gene expression profiling using exfoliated colonocytes: a pilot study. Biomarkers 8: 51-61. [Crossref]
- Ahmed FE, Ahmed NC, Vos P, Bonnerup C, Atkins JN, et al. (2012) Diagnostic microRNA markers to screen for sporadic human colon cancer in blood. Cancer Genom Proteom 9: 179-192. [Crossref]
- Abu Al-Soud W, Râdström P (1998) Capacity of nine thermostable DNA polymerases To mediate DNA amplification in the presence of PCR-inhibiting samples.Appl Environ Microbiol64: 3748-3753. [Crossref]
- Wilson IG (1997) Inhibition and facilitation of nucleic acid amplification.Appl Environ Microbiol63: 3741-3751. [Crossref]
- Rådström P, Knutsson R, Wolffs P, Lövenklev M, Löfström C (2004) Pre-PCR processing: strategies to generate PCR-compatible samples.Mol Biotechnol26: 133-146. [Crossref]
- Montiero I, Bonnemaison D, Vekris A, Petry KG, Bonnet J, et al. (1997) Comples polysaccharides as PCR inhibitors in feces: Heliobacter pylori model. J Clin Microbiol 104: 707-713.
- Schrader C, Schielke A, Ellerbroek L, Johne R (2012) PCR inhibitors- occurrence, properties and removal. J Appl Microbiol 113: 1014-1026.
- Scipioni A, Mauroy A, Ziant D, Saegerman C, Thiry E (2008) A SYBR Green RT-PCR assay in single tube to detect human and bovine noroviruses and control for inhibition.Virol J5: 94. [Crossref]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, et al. (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans.Nature403: 901-906. [Crossref]
- Xu P, Guo M, Hay BA (2004) MicroRNAs and the regulation of cell death.Trends Genet20: 617-624. [Crossref]
- Malumbres M (2013) miRNAs and cancer: an epigenetics view.Mol Aspects Med34: 863-874. [Crossref]
- Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, et al. (2007) Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2.Cell129: 303-317. [Crossref]
- Phua YL, Chu JY, Marrone AK, Bodnar AJ, Sims-Lucas S, et al. (2015) Renal stromal miRNAs are required for normal nephrogenesis and glomerular mesangial survival. Physiol Reports 3: e12537. [Crossref]
- Maes OC, Chertkow HM, Wang E, Schipper HM (2009) MicroRNA: Implications for Alzheimer Disease and other Human CNS Disorders.Curr Genomics10: 154-168. [Crossref]
- Li J, Li J, Liu X, Qin S, Guan Y, et al. (2013) MicroRNA expression profile and functional analysis reveal that miR-382 is a critical novel gene of alcohol addiction. EMBO Mol Med 5: 1402–1414. [Crossref]
- Romao JM, Jin W, Dodson MV, Hausman GJ, Moore SS, et al. (2011) MicroRNA regulation in mammalian adipogenesis.Exp Biol Med (Maywood)236: 997-1004. [Crossref]
- Mencía A, Modamio-Høybjør S, Redshaw N, Morín M, Mayo-Merino F, et al. (2009) Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss.Nat Genet41: 609-613. [Crossref]
- Hughes AE, Bradley DT, Campbell M, Lechner J, Dash DP, et al. (2011) Mutation altering the miR-184 seed region causes familial keratoconus with cataract.Am J Hum Genet89: 628-633. [Crossref]
- de Pontual L, Yao E, Callier P, Faivre L, Drouin V, et al. (2011) Germline deletion of the miR-17∼92 cluster causes skeletal and growth defects in humans.Nat Genet43: 1026-1030. [Crossref]
- Tuddenham L, Jung JS, Chane-Woon-Ming B, Dölken L, Pfeffer S (2012) Small RNA deep sequencing identifies microRNAs and other small noncoding RNAs from human herpesvirus 6B. J Virol 86: 1638–1649. [Crossref]
- Lu M, Zhang Q, Deng M, Miao J, Guo Y, et al. (2008) An analysis of human microRNA and disease associations.PLoS One3: e3420. [Crossref]
- Gregory RI, Shiekhattar R (2005) MicroRNA biogenesis and cancer.Cancer Res65: 3509-3512. [Crossref]
- Cummins JM, Velculescu VE (2006) Implications of micro-RNA profiling for cancer diagnosis.Oncogene25: 6220-6227. [Crossref]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, et al. (2005) A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia.N Engl J Med353: 1793-1801. [Crossref]
- Chen CZ (2005) MicroRNAs as oncogenes and tumor suppressors.N Engl J Med353: 1768-1771. [Crossref]
- Calin GA, Sevignai C, Dumitru CD, Hyslop T, Noch E, et al. (2004) Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 101: 2999-3004. [Crossref]
- Schepeler T, Reinert JT, Ostenfeld MS, Christensen LL, Silahtaroglu AN, et al. (2008) Diagnostic and prognostic microRNAs in stage II colon cancer.Cancer Res68: 6416-6424. [Crossref]
- Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, et al. (2008) MicroRNA expression profile associated with progression and therapeutic outcome in colon adenocarcinoma. J Am Med Assoc 299: 425-436.
- Calin GA, Croce CM (2006) MicroRNA signatures in human cancers.Nat Rev Cancer6: 857-866. [Crossref]
- Ansevin AT, Vizard DL, Brown BW, McConathy J (1976) High-resolution thermal denaturation of DNA. I. Theoretical & practical considerations for the resolution of thermal subtransitions. Biopolymers 15: 153-174. [Crossref]
- Ririe KM, Rasmussen RP, Wittwer CT (1997) Product differentiation by analysis of DNA melting curves during the polymerase chain reaction.Anal Biochem245: 154-160. [Crossref]
- Bruzzone CM, Tawadros PS, Boardman LA, Steer CJ (2013) Enhsanced primer selection and synthetic amplicon tempelates optimize high resolution melting analysis of single-nucleotide polymorphism in a large population. Genet Test Mol Biomarkers 17: 675-680. [Crossref]
- Mendes RE, Kiyota KA, Monteiro J, Castanheira M (2007) Rapid detection and identification of metallo-ß-lactamase encoding genes by multiplex -real-time PCR assay and melt curve analysis. J Clin Microbiol 45: 544-547.
- Guion CE, Ochoa TJ, Walker CM, Barletta F, Cleary TG (2008) Detection of diarrheagenic Escherichia coli by use of melting-curve analysis and real-time multiplex PCR.J Clin Microbiol46: 1752-1757. [Crossref]
- Varga A, James A (2005) Detection and differentiation of Plum pox virus using real-time multiplex PCR with SYBR Green and melting curve analysis: a rapid method for stain typing. J Virol Methods 123: 213-220. [Crossref]
- Tsiatis AC, Norris-Kirby A, Rich RG, Hafez MJ, Gocke CD, et al. (2010) Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of Kras mutations. J Mol Diag 12: 427-432. [Crossref]
- Bohling SD, Wittwer CT, King TC, Elenitoba-Johnson KS (1999) Fluorescence melting curve analysis for the detection of the bcl-1/JH translocation in mantle cell lymphoma.Lab Invest79: 337-345. [Crossref]




