The collection of disciplines referred to as “-omics” generally includes genomics, proteomics, and metabolomics, with other sub-fields including transcriptomics, metallomics, and lipidomics playing important roles in the context of emerging healthcare applications. The rapid development in computing power and artificial intelligence for the analysis of immense datasets has great potential for generating disease predicting models and may revolutionize personalized and preventative medicine in the near future. However, these algorithms rely on consistent formatting of data, turning unstructured clinical data into semi-structured data, and reliably annotating putative structures from mass spectrometry data, which are significant challenges. In addition, the complexity of biological systems complicates modeling outside of specific metabolic pathways or genetic interactions but hidden within this complexity are the biochemical fingerprints that can enable personalized treatment. Indeed, combining analyses of the -omics fields along the structure of the central dogma of biology would represents a herculean task, but would yield incredible predictive power. This review will introduce the major -omics fields listed above, introduced preliminary successes of these methods, and discuss their potential for application in biomedicine.
systems biology, metabolomics, proteomics, genomics, mass spectrometry, metallomics, next generation sequencing, AI, big data, machine learning
Next-generation personalized medicine has the potential to transform healthcare as we know it by considering individual differences in genetics, through genomics, and responses to environmental exposure, through metabolomics, lipidomics, and metallomics. The central dogma of biology describes the process of gene transcription and translation that produces proteins that act a biological catalysts and key gatekeepers of metabolic pathways (Figure 1). Systems biology seeks to analyze key players in this dogma, from gene to metabolite, to uncover novel associations, biomarkers of disease, and develop novel strategies for the generation of therapeutics, antibiotics, and antitumor drugs. This review will focus on recent developments in analytical chemistry and molecular biology that have led to the generation of a vast amount of “-omics” data that must be standardized and combined for a complete systems biology description of healthy and diseased states. While still in their infancy, the -omics fields have great potential for future healthcare applications, enabled by the power of big data and artificial intelligence (AI)-based solutions including machine learning.
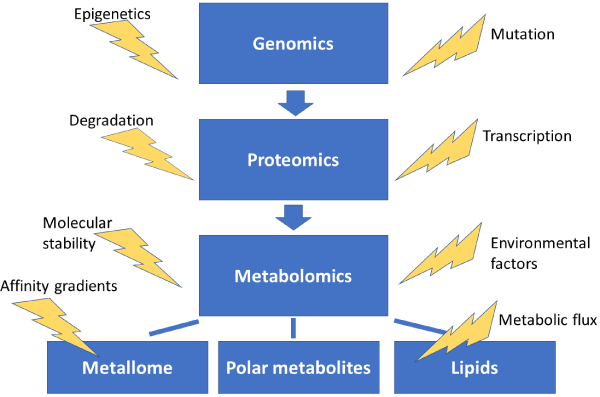
Figure 1. The central dogma of systems biology. The -omics fields and sub-fields are provided in the blue boxes with indicated influencing factors nearby. Because all fields and factors listed above can influence one another, the simplistic arrows provide only a general scheme of cascading influence. It should be noted that environmentally induced changes in the metabolome can result in a feedback loop, ultimately influencing genetics and gene products for example
Since the discovery of the DNA double helical structure, the chemical means of storing information, the field of genetics has expanded exponentially. The traditional model of Mendelian genetics and the “one-gene one-polypeptide” hypothesis have been demonstrated to be overly simplistic descriptions of the complexity of genetic interactions and expression. Indeed, spliceosomes can generate multiple products from a single gene, representing a significant source of genetic diversity that can be clinically relevant for analysis of certain cancers [1,2]. Thus, for accurate prediction of treatment outcomes and disease risks, both genotypic and phenotypic considerations must be incorporated for a personalized yet holistic assessment.
As the first -omics ¬ field, genomics was enabled by the development of DNA amplification techniques and automated sequencing in the 1980s and 90s [3-5]. The human genome project soon followed and promised unprecedent insight into various genetic diseases and ushered in the age of omics. Despite the successful mapping of the human genome [6], as well as those of other organisms [7], challenges remain for making sense of the immense amount of data, the functionality of non-coding regions [8], as well as genetic responses to environmental conditions [9].
Next-generation sequencing technologies have dramatically reduced the costs associated with genetic analysis, allowing for comparative studies that are important for evolutionary biology and analysis of polymorphisms on a large scale. This is essential for omics, as large datasets must be generated to build algorithms that can be used for various applications such as in silico drug repurposing for cancer patients [10]. The ability to inexpensively generate large amounts of meaningful data that is accurately annotated and deposited in accessible databases will be key to future -omics technologies. Thus the next-generation sequencing technologies have underpinned the explosion of potential for genomics to assist in the development of novel treatments and personalized medicine facilitated by individual genome sequencing [11,12].
One limitation of genomics is that the presence of a gene does not necessarily indicate its influence on the phenotype, which can be affected by protein synthesis and degradation rates, RNA splicing and silencing, and environment influences on the gene products. Thus, for a closer examination of phenotypes, examination of the functional products of genes, proteins, must be performed.
The field of proteomics has traditionally relied on chromatography coupled to mass spectrometry for data generation, but NMR studies have also made valuable contributions to the field. The power of high-resolution mass spectrometry can distinguish between proteins and various post-translational modifications (PTMs), which are often essential for protein function and serve as a key biological regulation process. For example, lysine acetylation was demonstrated to play a key role in regulating cellular metabolism through proteomic methods [13]. Thus, the genes coding for these proteins may not influence the predicted phenotype without post-translational activation by acetylation/phosphorylation or by association with co-factors, such as metals or small organic molecules (see the metallomics section).
As a representative example, Choudhary, et al. [14] used LC-MS/MS to identify 3600 lysine sites on 1750 proteins. From the massive dataset, the authors quantitatively assessed the acetylome and connected functionally related proteins. Such studies are necessary to unravel complex protein-protein interactions (often referred to as the interactome) and PTMs for a more complete understanding of systems biology. This is especially important as protein degradation can be signaled by PTMs and changes in the modification rates can manifest in altered protein concentrations and metabolic flux [15].
In addition to proteins, their substrates can be systematically examined to reveal various medical conditions and are the most sensitive to environmental factors and the most downstream target that are affected by protein activity and metabolic fluxes. The concentration and nature of metabolites is often thought of as the most sensitive phenotypic marker. In addition, the presence of exogenous metabolites may be indicative of infection and serve as a biomarker for clinical diagnoses. The metabolome is composed of many classes of compounds with diverse properties, making global analyses of all metabolites in a given system challenging. The metabolome is often crudely separated by polarity due to chromatographic considerations (see the Methodology section), with the hydrophobic “lipidome” considered a sub-field due to immense amount of structures that are classified as lipids.
Because of the diverse range of lipid classes and their individual importance in biological systems, the field of lipidomics is often distinguished from metabolomics [16] Since the mid-2000s, the rapid development in analytical equipment capable of separating (chromatography), detecting (mass spectrometry), and analyzing the structure of lipids (tandem mass spectrometry) has enabled large-scale data generation comparable to and even exceeding genomics and proteomics. The development of general (LipidBLAST and LipidPioneer) [17,18] and species-specific (MtB LipidDB) [19] lipid databases has facilitated rapid annotation and analysis via LC-MS/MS. In addition to the retention time, mass information, and characteristic MS/MS fragmentation patterns used to populate these databases, ion-mobility data has the potential to provide another layer for positive structure identification in a field where authentic standards for the vast amount of molecules are not available [20].
Using the complied and publicly-available databases, lipidomics has emerged as a promising option for the rapid identification of biomarkers of radiation exposure ovarian and prostate cancers, multiple sclerosis, nonalcoholic steatohepatitis and cardiovascular disease to name a few. For clinical application, however, challenges related to individual variation, sample preparation and handling, variability due to environmental factors, and instrumental accessibility must first be addressed [21-27].
As another sub-field of metabolomics, metallomics differs in that metal substrates are largely unchanged as they pass through metabolic pathways, unlike metabolites, and largely act as co-factors and structural components of proteins and enzymes. While the ligands and oxidation state of some metals can be altered, metal flux is largely governed by uptake, affinity gradients, equilibria, and export/import mechanisms [28].
Metal acquisition is an essential process for all organisms, and pathogenic bacteria have evolved specialized systems for obtaining metals such as iron, zinc, and copper from their hosts [29,30]. In response, hosts have development strict regulatory systems to limit the amount of “free” metals through a complex system of chaperones, transport proteins, storage proteins and high-affinity functional metalloenzymes [31-36].
In addition to isolated function studies of metalloproteins and determination of binding constants that drive the translocation of metals, whole cell extracts can be analyzed to determine the localization and importance of metals in a more biological context. Indeed, whole proteome analysis of free zinc levels and their responses to toxic metal exposure can reveal novel insights into detoxification mechanisms and metabolic responses to heavy metals [37,38]. This is often achieved using fluorescent metal sensors that are specific for certain metals and can be tuned in terms of binding affinity [39]. The Petering lab has pioneered the analysis of the Zn-proteome using reactions with various ligands and senors to classify biologically relevant zinc pools [40,41] In addition, the Lindahl group has identified iron species and the “ironome” response to various growth conditions in yeast and other cells with spatial resolution down to the mitochondria [42-44]. Understanding the speciation and roles that metals play in response to various conditions is key to a complete understanding of bioinorganic chemistry and its role in systems biology.
A Methodologies for next-generation -omics: mass spectrometry as a multi-functional analytical tool (Figure 2)
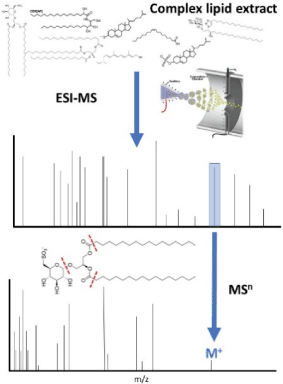
Figure 2. An example of lipidomic profiling using tandem MS (MSn), which can be used reveal identities of key lipids through mass matching and fragmentation pattern assignment and provide a platform for quantification
Recent advances in analytical techniques have revolutionized the field of metabolite and lipid biology allowing an “-omics” approach to be taken to tackle tough questions regarding lipid structure and function [45]. The adaptability of MS allows it to act as a swiss army knife of analytical techniques with the ability to deliver both quantitative and qualitative information [46,47]. This emerging multi-functionality has yet to be fully exploited and holds great promise.
In addition, to probe global protein conformation, ion-mobility (IM)-MS is becoming an increasingly popular tool which can easily distinguish between conformers in solution by drift times in a gas flow, which are governed by the overall shape of the molecule [48-50]. IM-MS has also been applied to the determination of lipid structure and distinguish between proteins and isomers [51]. This provides a second level of molecular identification after the crude MS mass measurement and can facilitate more accurate identification of metabolites.
As an additional level of structure analysis that can be used for peak annotation, tandem MS (MSn) is commonly used in proteomics and metabolomic analysis and it fragments the protein or small molecule. The resulting fragmentation pattern can be analysed to determine structure and pinpoint modification or ligand binding sites [52-54]. MSn is needed to distinguish between different lipids with the same m/z and locate modified residues in proteins. Thus, the MS platform is multi-functional for dynamic -omic analysis of the interactions of lipids, metabolites, and proteins during disease and infection progression and can be used for detailed studies of virulence and health outcomes.
The discovery of novel virulence factors can be achieved using MS methods for high-throughput analysis of bacterial metabolites. Limitations due to the vast array of lipids and other metabolites present in an extract and unequal ionization efficiency can be overcome by lipid-class specific modification protocols that facilitate their analysis by MS [55]. In addition to qualitative data regarding the identity of lipids, polar metabolites, and proteins, quantitative analysis can be achieved through the use of isotopically labelled internal standards or control cell extracts grown in isotopically-enriched media [56].
The cornucopia of lipid species can be separated simply by switching ion modes in the MS.[57,58] Species such as wax diesters, saccharolipids and free fatty acids ionize readily in negative-ion mode whereas classes such as diacyltrehaloses, glycerolipids and mycobactins are found primarily in the positive-ion spectra [59,60]. Further separation can be achieved through chromatographic and ion-mobility methods in addition to MSn, which can distinguish between different lipid structures with the same mass [61].
By analysing vital lipid concentrations during the infection process, key insights can be gained into their function and infection mechanisms [62]. In addition, studying the lipid composition of gene knockouts will elucidate bacterial strategies for metabolism modification in response to shutdown of important biochemical pathway by antibiotics [63,64] Upon identification of backup metabolic pathways, synergistic antibiotic intervention strategies can be tested where multiple pathways are targeted to prevent the development of resistance and make the overall treatment more effective [65]. By taking a systems approach to tackle antibiotic-resistant infections, more effective interventions can be developed to improve patient outcomes. Thus, MS-based metabolomic approaches have significant potential applications in screening, environmental monitoring, fundamental biological studies, and rapid identification of virulent strains of common pathogens.
In the field of metallomics, inductively coupled plasma (ICP)-MS is useful for studying metal distribution and associate species after chromatographic separation or tissue fixation [66,67]. In addition, techniques such as Mossbauer spectroscopy and EXAFS can be used to probe metal species distribution in biological samples, although they typically require significant sample preparation [68].
The significant amount of data generated by the previously described -omics fields provides a foundation that may allow future researchers to “stand on the shoulders of giants”, which may be more accurately be referred to standing on the shoulders of giant databases. Through mining of the significant amount of data generation, correlations can be drawn between seemingly disparate experiments and datasets to more clearly understand medically and biologically important phenomena [69].
In addition, significant amounts of data from electronic medical records which have the potential to be analysed for the discovery of novel drug-gene interactions and a refined characterization of pharmaceutical side-effects [70]. This, big data will further enable systems and clinical pharmacological approaches at an integrative and holistic level that can be applied to an individual for patient-specific treatment and healthcare maintenance.
Gene variations play an important role in determining a person’s response to medication, affecting the medication’s efficacy and/or safety profile [71]. While each person’s genetic makeup is unique, most modern day medications are developed in a “one-size-fits-all” manner, leading to sub-optimal treatment outcomes. Pharmacogenetics enhance patient care by combining genomics and pharmacology to customize patient-specific treatments which lower adverse drug event (ADE) risks and optimize dosing. Additionally, pharmacogenomics can proactively distinguish drug responders from non-responders to further assist in selecting the most ideal therapy for a patient [72]. The understanding of the genetic foundation and inter-patient variability in drug responses have propelled clinical medicine towards individualized patient care management. Pharmacokinetics play a key role in pharmacogenomics, which studies genetic variations in enzymes, receptors, and transporters which affect a drug’s metabolism, elimination, absorption, and distribution [73].
Pharmacogenomics have been appreciated in the many specialty medical fields. Substantial advances in genome interrogation techniques have identified pharmacogenomic biomarkers for several cardiovascular drugs including: clopidogrel, warfarin, and beta-blocking agents. For example, clopidogrel is an antiplatelet agent by inhibiting platelet P2Y12 receptors and is used to prevent stent thrombosis in patients who require stent placement after an acute coronary syndrome to decrease mortality rates [74]. Clopidogrel is an inactive prodrug that must be metabolized to its bioactive form by the hepatic enzyme protein CYP2C19. Because of 25 known polymorphic gene variation of CYP2C19, patient response to this therapy varies greatly depending on individual genetics. Genotyping CYP2C19 allow clinicians to identify patients who would be poor responders or potentially resistant to clopidogrel, and guide them towards more effective antiplatelet therapies.
Warfarin an anticoagulation agent used to treat and prevent venous thromboembolism as well as to prevent stroke in patients with atrial fibrillation. Warfarin targets and inhibits the enzyme vitamin K epoxide reductase complex subunit-1 (VKORC1) to prevent the activation of clotting factors VII, IX, X, II, and proteins C and S [75]. Warfarin is metabolized by hepatic enzyme protein CYP2C9. Variations in VKORC1 and CYP2C9 play a role in variation in warfarin responses and are genetic determinants of dosing [76]. A marker of warfarin resistance is the VKORC1 D36Y variant, which can be identified by pharmacogenomic advancements and technology.
Beta-blocking agents inhibit the beta-1 adrenoreceptors and decrease catecholamine stimulation. This class of medication shows improved mortality in patients with heart failure by preventing and reversing cardiac remodeling. Beta-blockers are metabolized by the enzyme CYP2D6 and some individual carry a variant of the CYP2D6 enzyme that decreases their ability to metabolize this medication, thereby increasing the risk of symptomatic hypotension and bradycardia [77]. While patients are not readily tested for the gene variation, pharmacogenomics played a clinical role in beta-blocker treatments in patients. The Federal Drug Association (FDA) utilized pharmacogenomics to identify that 8% of the Caucasian population lack this enzyme altogether. The use of pharmacogenomics allows clinicians to identify patients who are on concomitant medications that inhibit the CYP2D6 enzyme and are at higher risk of being poor metabolizers of beta-blockers. In doing so, healthcare providers can provide the most optimal care for their patients who require this class of medication.
Substantial advancements in the field of pharmacogenomics have been recognized by the FDA. Biomarkers in FDA drug labeling include the specific genetic biomarkers and their role in drug exposure which affects response variability and risks of adverse drug events and the inclusion of pharmacogenomics in drug labeling have assisted providers in drug selection and dosing recommendations which are optimal for their specific patients.
While still in its infancy, pharmacogenomics have been useful in precision medicine and individualizing patient care. However, its application is not readily accessible nor affordable, thus not a viable option for many patients and providers. Further advancements are required before this technique is readily utilized in normal practice, but with the development of big data and AI technologies the field may becoming increasingly accessible in the coming years.
The powerful analytical tools, some of which were described herein, used in the -omics fields have generated massive amounts of data and provided unprecedented insight into various biological process. Future challenges will include harmonization of disparate datasets containing various types of information from the various -omics fields and subsequent algorithmic analysis to identify novel interactions, drug uses, and predict patient responses to certain treatments. While still in its early stages, the -omics fields exhibit significant promise as medicine shifts towards a broader focus of the individual as a dynamic system at the confluence of genes, proteins, metabolites, and metals.
- Worby CA, Simonson-Leff N, Clemens JC, Kruger RP, Muda M, et al. (2001) The sorting nexin, DSH3PX1, connects the axonal guidance receptor, Dscam, to the actin cytoskeleton. J Biol Chem 276: 41782-41789. [Crossref]
- Gibb EA, Enfield KS, Tsui IF, Chari R, Lam S, et al. (2011) Deciphering squamous cell carcinoma using multidimensional genomic approaches. J Skin Cancer 2011: 541405. [Crossref]
- Mullis K, Faloona F, Scharf S, Saiki R, Horn G, et al. (1986) In Cold Spring Harbor symposia on quantitative biology; Cold Spring Harbor Laboratory Press: 51: 263.
- Smith LM, Sanders JZ, Kaiser RJ, Hughes P, Dodd C, et al. (1986) Fluorescence detection in automated DNA sequence analysis. Nature 321: 674-679. [Crossref]
- Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, et al. (1991) Complementary DNA sequencing: expressed sequence tags and human genome project. Science 252: 1651-1656. [Crossref]
- Istrail S, Sutton GG, Florea L, Halpern AL, Mobarry CM, et al. (2004) Whole-genome shotgun assembly and comparison of human genome assemblies. Proc Natl Acad Sci U S A 101: 1916-1921. [Crossref]
- Cheng S, Melkonian M, Smith SA4, Brockington S5, et al. (2018) 10KP: A phylodiverse genome sequencing plan. Gigascience 7: 1-9. [Crossref]
- Williams SM, An JY, et al. (2018) An integrative analysis of non-coding regulatory DNA variations associated with autism spectrum disorder. Mol Psychiatry. [Crossref]
- Baker BH, Berg LJ, Sultan SE (2018) Context-Dependent Developmental Effects of Parental Shade Versus Sun Are Mediated by DNA Methylation. Front Plant Sci 9: 1251. [Crossref]
- Cheng F, Lu W, Liu C, Fang J, Hou Y, et al. (2019) Nature communications.
- Besser J, Carleton HA, Gerner-Smidt P, Lindsey RL, Trees E (2018) Next-generation sequencing technologies and their application to the study and control of bacterial infections. Clin Microbiol Infect 24: 335-341. [Crossref]
- Nagahashi M, Shimada Y, Ichikawa H, Kameyama H, Takabe K, et al. (2019) Next generation sequencing-based gene panel tests for the management of solid tumors. Cancer sci 110: 6-15. [Crossref]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, et al. (2010) Regulation of cellular metabolism by protein lysine acetylation. Science 327: 1000-1004. [Crossref]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, et al. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325: 834-840. [Crossref]
- Wu X, Gong F, Cao D, Hu X, Wang W (2016) Proteomics. 16: 847.
- Wenk MR (2005) The emerging field of lipidomics. Nat Rev Drug Discov 4: 594-610. [Crossref]
- Cajka T, Fiehn O (2017) LC-MS-Based Lipidomics and Automated Identification of Lipids Using the LipidBlast In-Silico MS/MS Library. Methods Mol Biol 1609: 149-170. [Crossref]
- Ulmer CZ, Koelmel JP, Ragland JM, Garrett TJ, Bowden JA (2017) LipidPioneer: A Comprehensive User-Generated Exact Mass Template for Lipidomics. J Am Soc Mass Spectrom 28: 562-565. [Crossref]
- Sartain MJ, Dick DL, Rithner CD, Crick DC, Belisle JT (2011) Lipidomic analyses of Mycobacterium tuberculosis based on accurate mass measurements and the novel "Mtb LipidDB". J Lipid Res 52: 861-872. [Crossref]
- Paglia G, Astarita G (2017) Metabolomics and lipidomics using traveling-wave ion mobility mass spectrometry. Nat Protoc 12: 797-813. [Crossref]
- Pannkuk EL, Laiakis EC, Mak TD, Astarita G, Authier S, et al. (2016) A Lipidomic and Metabolomic Serum Signature from Nonhuman Primates Exposed to Ionizing Radiation. Metabolomics p. 12. [Crossref]
- Zhang Y, Liu Y, Li L, Wei J, Xiong S, et al. (2016) High resolution mass spectrometry coupled with multivariate data analysis revealing plasma lipidomic alteration in ovarian cancer in Asian women. Talanta 150: 88-96. [Crossref]
- Zhou X, Mao J, Ai J, Deng Y, Roth MR, et al. (2012) Identification of plasma lipid biomarkers for prostate cancer by lipidomics and bioinformatics. PLoS One 7: e48889. [Crossref]
- Del Boccio P, Pieragostino D, Di Ioia M, Petrucci F, Lugaresi A, et al. (2011) Lipidomic investigations for the characterization of circulating serum lipids in multiple sclerosis. J Proteomics 74: 2826-2836. [Crossref]
- Loomba R, Quehenberger O, Armando A, Dennis EA (2015) Polyunsaturated fatty acid metabolites as novel lipidomic biomarkers for noninvasive diagnosis of nonalcoholic steatohepatitis. J Lipid Res 56: 185-192. [Crossref]
- Hinterwirth H, Stegemann C, Mayr M (2014) Lipidomics: quest for molecular lipid biomarkers in cardiovascular disease. Circ Cardiovasc Genet 7: 941-954.
- Rai S, Bhatnagar S (2017) Novel Lipidomic Biomarkers in Hyperlipidemia and Cardiovascular Diseases: An Integrative Biology Analysis. OMICS 21: 132-142. [Crossref]
- Banci L, Bertini I, Ciofi-Baffoni S, Kozyreva T, Zovo K, et al. (2010) Affinity gradients drive copper to cellular destinations. Nature 465: 645-648. [Crossref]
- Tiedemann MT, Pinter TB, Stillman MJ (2012) Insight into blocking heme transfer by exploiting molecular interactions in the core Isd heme transporters IsdA-NEAT, IsdC-NEAT, and IsdE of Staphylococcus aureus. Metallomics 4: 751-760. [Crossref]
- Tiedemann MT, Heinrichs DE, Stillman MJ (2012) Multiprotein heme shuttle pathway in Staphylococcus aureus: iron-regulated surface determinant cog-wheel kinetics. J Am Chem Soc 134: 16578-16585. [Crossref]
- Adlard PA, Parncutt J, Lal V, James S, Hare D, et al. (2015) Metal chaperones prevent zinc-mediated cognitive decline. Neurobiol Dis 81: 196-202. [Crossref]
- Taylor JM, Heinrichs DE (2002) Transferrin binding in Staphylococcus aureus: involvement of a cell wall-anchored protein. Mol Microbiol 43: 1603-1614. [Crossref]
- Ott DB, Hartwig A, Stillman MJ (2019) Competition between Al3+ and Fe3+ binding to human transferrin and toxicological implications: structural investigations using ultra-high resolution ESI MS and CD spectroscopy. Metallomics 11: 968-981. [Crossref]
- Orihuela R, Fernández B, Palacios O, Valero E, Atrian S, et al. (2011) Ferritin and metallothionein: dangerous liaisons. Chem Commun (Camb) 47: 12155-12157. [Crossref]
- Pinter TB, Stillman MJ (2014) The zinc balance: competitive zinc metalation of carbonic anhydrase and metallothionein 1A. Biochemistry 53: 6276-6285. [Crossref]
- Pinter TB, Irvine GW, Stillman MJ (2015) Domain Selection in Metallothionein 1A: Affinity-Controlled Mechanisms of Zinc Binding and Cadmium Exchange. Biochemistry 54: 5006-5016. [Crossref]
- Namdarghanbari MA, Bertling J, Krezoski S, Petering DH (2014) Toxic metal proteomics: reaction of the mammalian zinc proteome with Cd²âº. J Inorg Biochem 136: 115-121. [Crossref]
- Petering DH (2016) Reactions of the Zn Proteome with Cd2+ and Other Xenobiotics: Trafficking and Toxicity Chemical research in toxicology Chem Res Toxicol 30: 189-202.
- Karim MR, Petering DH (2016) Newport Green, a fluorescent sensor of weakly bound cellular Zn(2+): competition with proteome for Zn(2). Metallomics 8: 201-210. [Crossref]
- Nowakowski AB, Petering DH (2011) Reactions of the fluorescent sensor, Zinquin, with the zinc-proteome: adduct formation and ligand substitution. Inorg Chem 50: 10124-10133. [Crossref]
- Meeusen JW, Nowakowski A, Petering DH (2012) Reaction of metal-binding ligands with the zinc proteome: zinc sensors and N,N,N',N'-tetrakis(2-pyridylmethyl)ethylenediamine. Inorg Chem 51: 3625-3632. [Crossref]
- Holmes-Hampton GP, Jhurry ND, McCormick SP, Lindahl PA (2013) Iron content of Saccharomyces cerevisiae cells grown under iron-deficient and iron-overload conditions. Biochemistry 52: 105-114. [Crossref]
- Jhurry ND, Chakrabarti M, McCormick SP, Holmes-Hampton GP, Lindahl PA (2012) Biophysical investigation of the ironome of human jurkat cells and mitochondria. Biochemistry 51: 5276-5284. [Crossref]
- McCormick SP, Moore MJ, Lindahl PA (2015) Detection of Labile Low-Molecular-Mass Transition Metal Complexes in Mitochondria. Biochemistry 54: 3442-3453. [Crossref]
- Triebl A, Trötzmüller M, Hartler J, Stojakovic T, Köfeler HC (2017) Lipidomics by ultrahigh performance liquid chromatography-high resolution mass spectrometry and its application to complex biological samples. J Chromatogr B Analyt Technol Biomed Life Sci 1053: 72-80. [Crossref]
- Wang M, Wang C, Han X (2016) Selection of internal standards for accurate quantification of complex lipid species in biological extracts by electrospray ionization mass spectrometry-What, how and why? Mass spectrom rev 6: 693-714 [Crossref]
- Garrett TA (2016) Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids.
- Lanucara F, Holman SW, Gray CJ2, Eyers CE1 (2014) The power of ion mobility-mass spectrometry for structural characterization and the study of conformational dynamics. Nat Chem 6: 281-294. [Crossref]
- Zhou M, Politis A, Davies R, Liko I, et al. (2014) Ion mobility-mass spectrometry of a rotary ATPase reveals ATP-induced reduction in conformational flexibility. Nat Chem 6: 208-215. [Crossref]
- Chen SH1, Chen L, Russell DH (2014) Metal-induced conformational changes of human metallothionein-2A: a combined theoretical and experimental study of metal-free and partially metalated intermediates. J Am Chem Soc 136: 9499-9508. [Crossref]
- Hines KM, May JC, McLean JA, Xu L (2016) Evaluation of Collision Cross Section Calibrants for Structural Analysis of Lipids by Traveling Wave Ion Mobility-Mass Spectrometry. Anal Chem 88: 7329-7336. [Crossref]
- Han X, Gross RW (1995) Structural determination of picomole amounts of phospholipids via electrospray ionization tandem mass spectrometry. J Am Soc Mass Spectrom 6: 1202-1210. [Crossref]
- Witze ES, Old WM, Resing KA, Ahn NG (2007) Mapping protein post-translational modifications with mass spectrometry. Nat Methods 4: 798-806. [Crossref]
- Li H, Wongkongkathep P, Van Orden SL, Ogorzalek Loo RR, Loo JA (2014) Revealing ligand binding sites and quantifying subunit variants of noncovalent protein complexes in a single native top-down FTICR MS experiment. J Am Soc Mass Spectrom 25: 2060-2068. [Crossref]
- Tsikas D, Zoerner AA, Mitschke A, Gutzki FM (2009) Nitro-fatty acids occur in human plasma in the picomolar range: a targeted nitro-lipidomics GC-MS/MS study. Lipids 44: 855-865. [Crossref]
- Yang K, Han X (2011) Accurate quantification of lipid species by electrospray ionization mass spectrometry - Meet a key challenge in lipidomics. Metabolites 1: 21-40. [Crossref]
- Schuhmann K, Almeida R, Baumert M, Herzog R, Bornstein SR, et al. (2012) Shotgun lipidomics on a LTQ Orbitrap mass spectrometer by successive switching between acquisition polarity modes. J Mass Spectrom 47: 96-104. [Crossref]
- Breitkopf SB, Ricoult SJ, Yuan M, Xu Y, Peake DA, et al. (2017) A relative quantitative positive/negative ion switching method for untargeted lipidomics via high resolution LC-MS/MS from any biological source. Metabolomics 13: 30. [Crossref]
- Ellis SR, Soltwisch J, Paine MRL, Dreisewerd K, Heeren RMA (2017) Laser post-ionisation combined with a high resolving power orbitrap mass spectrometer for enhanced MALDI-MS imaging of lipids. Chem Commun (Camb) 53: 7246-7249. [Crossref]
- Ovcacíková M, Lísa M, Cífková E, Holcapek M (2016) Retention behavior of lipids in reversed-phase ultrahigh-performance liquid chromatography–electrospray ionization mass spectrometry. J Chromatogr A 1450: 76-85.
- Baker PR, Armando AM, Campbell JL, Quehenberger O, Dennis EA (2014) Three-dimensional enhanced lipidomics analysis combining UPLC, differential ion mobility spectrometry, and mass spectrometric separation strategies. J Lipid Res 55: 2432-2442. [Crossref]
- Jain M, Petzold CJ, Schelle MW, Leavell MD, Mougous JD, et al. (2007) Lipidomics reveals control of Mycobacterium tuberculosis virulence lipids via metabolic coupling. Proc Natl Acad Sci U S A 104: 5133-5138. [Crossref]
- Lahiri N, Shah RR, Layre E, Young D, Ford C, et al. (2016) Rifampin Resistance Mutations Are Associated with Broad Chemical Remodeling of Mycobacterium tuberculosis. J Biol Chem 291: 14248-14256. [Crossref]
- Mendum TA, Wu H, Kierzek AM, Stewart GR (2015) Lipid metabolism and Type VII secretion systems dominate the genome scale virulence profile of Mycobacterium tuberculosis in human dendritic cells. BMC Genomics 16: 372. [Crossref]
- Fügi MA, Kaiser M, Tanner M, Schneiter R, Mäser P, et al. (2015) Match-making for posaconazole through systems thinking. Trends Parasitol 31: 46-51. [Crossref]
- Becker JS, Matusch A, Palm C, Salber D, Morton KA, et al. (2010) Bioimaging of metals in brain tissue by laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS) and metallomics. Metallomics 2: 104-111. [Crossref]
- Vogiatzis CG, Zachariadis GA (2014) Tandem mass spectrometry in metallomics and the involving role of ICP-MS detection: a review. Anal Chim Acta 819: 1-14. [Crossref]
- Wofford JD, Chakrabarti M, Lindahl PA (2017) Mössbauer Spectra of Mouse Hearts Reveal Age-dependent Changes in Mitochondrial and Ferritin Iron Levels. J Biol Chem 292: 5546-5554. [Crossref]
- Mullins IM, Siadaty MS, Lyman J, Scully K, Garrett CT, et al. (2006) Data mining and clinical data repositories: Insights from a 667,000 patient data set. Comput Biol Med 36: 1351-1377. [Crossref]
- Jensen PB, Jensen LJ, Brunak S (2012) Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet 13: 395-405. [Crossref]
- Relling MV, Evans WE (2015) Pharmacogenomics in the clinic. Nature 526: 343-350. [Crossref]
- Johnson JA, Cavallari LH, Beitelshees AL, Lewis JP, Shuldiner AR, et al. (2011) Pharmacogenomics: Application to the Management of Cardiovascular Disease. Clin Pharmacol Ther 90: 519.
- Crews KR, Hicks JK, Pui CH, Relling MV, Evans WE (2012) Pharmacogenomics and individualized medicine: Translating science into practice. Clin Pharmacol Ther 92: 467-475. [Crossref]
- Amsterdam EA, Wenger NK, Brindis RG, Casey DE, Ganiats TG, et al. (2014) 2014 AHA/ACC Guideline for the Management of Patients with Non-ST-Elevation Acute Coronary Syndromes: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol 64: e139-e228. [Crossref]
- Bell RG, Sadowski JA, Matschiner JT (1972) Mechanism of action of warfarin. Warfarin and metabolism of vitamin K. Biochemistry 11: 1959-1961. [Crossref]
- Wadelius M, Chen LY, Downes K, Ghori J, Hunt S, et al. (2005) Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogenomics J 5: 262-270. [Crossref]
- Rau T, Heide R, Bergmann K, Wuttke H, Werner U, et al. (2002) Effect of the CYP2D6 genotype on metoprolol metabolism persists during long-term treatment. Pharmacogenetics 12: 465-472. [Crossref]


