Abstract
Cell signal transduction is a complex system of communication that governs basic cellular activities and coordinates cell actions. Adaptor proteins containing SH2 or/and SH3 domain play critical roles in cell signaling and diseases. Recently, SH3 domain-binding glutamic acid-rich (SH3BGR) protein family members were characterized, except SH3BGRL3, they share a consensus SH3-binding motif and a Homer EVH1 binding motif, and function in physiological and pathological events of many diseases, including Down syndrome, diabetes and cancers. In this review, we summarized the recent advances in expression patterns and physiological roles of SH3BGR family members in model systems and cancers, and suggested the further endeavors to unmask their real facets in homeostasis and pathogenecity when dysfunctional to provide potential hints for the diagonosis and therapy of related diseases.
Key words
SH3BGR family, adaptor protein, expression pattern, physiology, cancer
Introduction
Cell signal conduction is a complex system of communications that governs basic cell physiologies and coordinates cellular activities. In this process, protein interactions allow extracellular signals to select the respective cell and plasma membrane receptors and subsequently to conduct information to specific intracellular sites, which is an essential facet of cellular regulation [1]. Protein interactions, especially those involved SH3 domains to recruit enzymes into signaling networks or place enzymes close to their substrates are commonly applied in signal transduction, cytoskeletal rearrangements, membrane trafficking, and other key cellular regulations [2,3]. Recently, a SH3 domain-binding glutamic acid-rich (SH3BGR) protein family composed of SH3BGR [4], SH3BGRL [5], SH3BGRL2 [6] and SH3BGRL3 [7] is characterized by the presences of a proline-rich region with SH3-binding motif and an acidic carboxyl terminal region with Homer EVH1 binding motif, respectively [4]. The presence of two domains often involved in protein-protein interactions suggests that SH3BGR protein family may play multiple roles in signaling transduction processes of cells.
Biological characters of SH3BGR family
SH3 domain-binding glutamic acid rich (SH3BGR) was firstly found in the identification of genes involved in the pathogenesis of Down Syndrome patients and named based on two contained motifs, the proline-rich region and the glutamic acid rich region [4]. After characterization of other homologs, including SH3BGRL [5], SH3BGL2 [6] and SH3BGRL3 [7], the shared highly conserved N-terminal region with a proline-rich sequence (PLPPQIF), a SH3 binding (PXXP) [1] and the Homer EVH1 binding (PPXXF) motifs [1,8] were featured, respectively (Figure 1), thus these four genes were assigned as a new SH3BGR gene family [6].
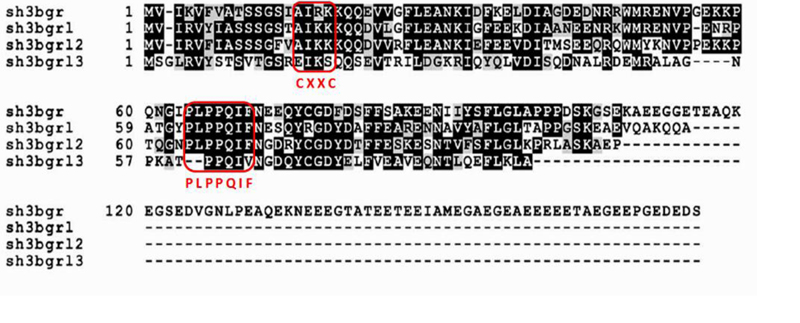
Figure 1. ClustalW alignment of the amino acid sequences of the human SH3BGRL family.
Identical and conserved residues are shaded in black and gray, respectively. The conservative proline-rich motif PLPPQIF and the lacking catalytic motif Cys-X-X-Cys in the human SH3BGR family are boxed in red. This figure is cited with modification from Ref. [6].
Although SH3BGR family members have very similar structure, their loci in genome are distinct. sh3bgr is mapped to chromosome 21q22.3 [4], sh3bgrl, sh3bgrl2 and sh3bgrl3 locate in chromosome Xq13.3 [5], chromosome 6q13-15 [6] and chromosome 1p34.3-35 [7], respectively. The member sh3bgr encodes a 239 amino acids protein that contains a highly conserved N-terminal region of about 100 amino acids and a variable C-terminal region highly enriched with glutamic acid residues [4]. sh3bgrl encodes a small protein of 114 amino acids, sharing 60% identical and 84% conservative amino acids with the middle portion, proline-rich region of the sh3bgr gene [5]. sh3bgrl2 encodes a protein of 107 amino acids and appears to be related to Glutaredoxins and PKC-interacting cousin of thioredoxin (PICOT) homology domain [6]. sh3bgrl3 encodes a small protein of 93 amino acids, showing 39% identity and 76% similarity to SH3BGR protein, but lacks both SH3 and Homer EVH1 binding motifs, indicating that SH3BGRL3 may function differently from other members of the family [7]. Whatever, all SH3BGR members have very high similarity to Glutaredoxin1 (GRX1) of Escherichia coli at both amino acid sequence and the predicted structural level [7], but they completely lacks the CXXC motif that is essential for glutaredoxin enzymatic function [9]. Given the structural feature, it could be expected that SH3BGR members may participate the redox-dependent processes in cells [7], but there are no enough evidence to support this hypothesis.
Expression pattern, physiological and pathological functions of SH3BGR family
Since identification of SH3BGR family, their physiological functions in vivo are largely unknown. Expression patterns of SH3BGR and SH3BGRL in Xenopus have been reported and indicated that Sh3bgr could be critical for sarcomere formation in striated muscles through regulating localization of Enah, and heart development formation by affecting the Enah protein level in Xenopus [10]. Our current organ-specific expression analysis of SH3BGR members in zebrafish embryos demonstrated that sh3bgrl3 is uniquely expressed in liver, and sh3bgr in sarcomere (unpublished data). Similarly, the mRNA expression of all four members are also expressed in chicken and demonstrated that sh3bgr is restricted to muscle tissues, both sh3bgrl and sh3bgrl3 are highly expressed in lymphoid tissues (spleen, bursa, and thymus), exhibiting low levels of expression in the liver, skeletal muscle, and kidney, while sh3bgrl2 is expressed highest in the brain and lung, and marginally expressed in the other tissues [11].
So far, to our knowledge, no knockout mice model is reported to dissect the developmental and physiological functions of these genes in mammals. Considering that SH3BGR protein family was screened from the pathogenesis of congenital heart disease (CHD) in Down Syndrome (DS) [4] and Sh3bgr is strictly expressed in the earliest stages of mouse heart development [12], it is possible this gene family plays a role in heart morphogenesis and, consequently, in the pathogenesis of CHD in DS. When forcing expression of Sh3bgr in transgenic mice with a cardiac-and skeletal-muscle-specific promoter, it seems that SH3BGR has no effect on heart morphogenesis [13]. Moreover, SH3BGR was activated in STAT2 high-expression metabolism coupling postmitotic outgrowth to visual and sound perception network [14].
SH3BGRL was found to be one of 16 proteins differentially expressed in Parkinson disease (PD) patients comparing with healthy people [15]. For SH3BGRL2, although higher level of SH3BGRL2 expression was reported to participate in erythropoietin (EPO) stimulated erothrocyte differentiation [16], the expression level of SH3BGRL2 in peripheral blood lymphomononuclear cells (PBMC) showed no difference between Type I diabetes and Type II diabetes [17]. SH3BGRL3, previously named TIP-B1 (TNF inhibitory protein-B1) [18] was upregulated under TNF prior simulation to protect cells from TNF induced apoptosis [18,19]. In addition, higher expression of SH3BGRL3 in CD4+ T lymphocytes was proved to participate in the pathogenesis of systemic lupus erythematosus (SLE) [20]. We summarized all these milestone events of SH3BGR family from their first identification (Figure 2).
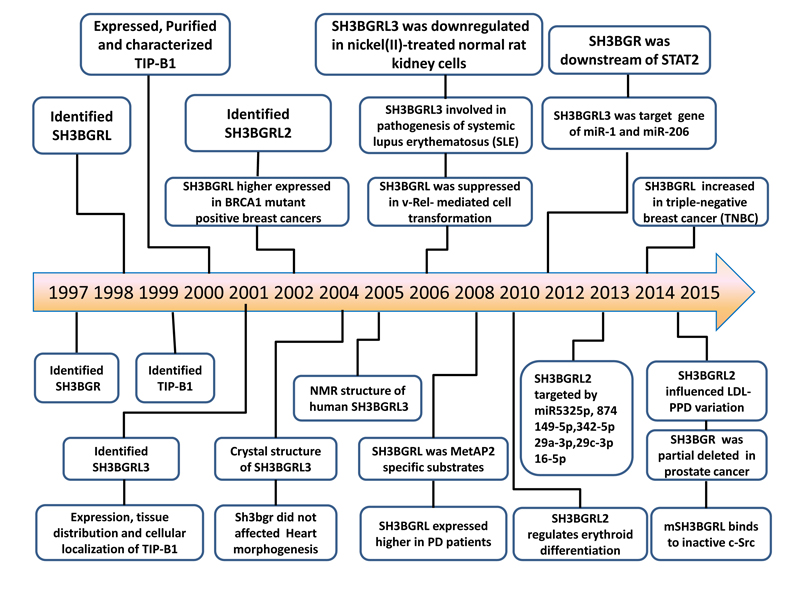
Figure 2. Major achievements in the research of SH3BGR family.
After SH3BGR and its homologs were identified in Down Syndrome (DS), a new family was established as a thioredoxin-like protein superfamily. The family members, except SH3BGRL3, share both functional SH3-binding and EVH1-binding motifs, which could be involved in various physiological and pathological processes, including in cancers.
Function of SH3BGR member in tumorigenesis
In cancers, SH3BGRL was demonstrated to be overexpressed in BRCA1 mutated breast tumors [21], ER positive breast tumors [22] and oral squamous carcinoma, even higher in the invasive oral squamous carcinomas and breast cancers, compared to the non-invasive counterparts or normal breast tissues [23]. Overexpression of murine SH3BGRL effectively promoted tumor formation and metastasis in xenagraft model [23]. Unexpectedly, human SH3BGRL is a tumor suppressor [23], which is consistent to the observation that SH3BGRL can repress the viral v-Rel-induced cell transformation [11]. Mechanistically, somatic mutation of human SH3BGRL, such as R76C could trigger tumorigenesis and pulmonary metastasis as murine SH3BGRL [23], which may partially interpret why human SH3BGRL is upregulated in various cancers. Moreover, Blocking c-Src and FAK attenuated the downstream AKT and MAPK activities, leading to much less tumor masses and metastatic sites, which provided a possible strategy for the related cancer therapy. However, based on the TCGA dataset, the somatic mutation frequency of human SH3BGRL is quite low, even less than 5%. Therefore, we suspect the protein modification could be involved in governing SH3BGRL protein stablility and its physiological functions,dependent of the particular cell contexts. Therefore, much effort should be placed on dissecting the detailed mechanisms how SH3BGR members work, what interacting partners are involved in both normal and transformed cells and tissues.
SH3BGRL3 was found to be downregulated in nickel (II)-induced carcinogenesis as well in normal rat kidney cells [24]. Further study uncovered that it can be targeted by small noncoding RNAs miR-1 and miR-206 to regulate C2C12 myoblast cell differentiation [25], suggesting the tumor-repressing role of SH3BGRL3. In contrast, SH3BGRL3 protein was recently observed to be upregulated in urothelial carcinoma samples, and molecularly binds to epidermal growth factor receptor for tumorigenesis, indicating that it could be a potential prognostic biomarker [26]. Similar to SH3BGRL, the real function of these adaptor proteins would be mainly dependent on the specific cell components, and their protein modification would also be a critical cause to their functions as well.
Except the above two players, neither SH3BGR nor SH3BGRL2 is yet to be documented in tumorigenesis. Thus the detailed function and the underlying mechanism of SH3BGR members need to be investigated, especially in animal tumor models.
Signaling pathways regulated by SH3BGR family
SH3BGRL proteins mainly locate in nucleus, only small portion in cytosol and on the inner cell membrane [23]. As a simple protein family with only a SH3 motif, a proline-rich region and a EVH1 domain, it is positively expected that SH3BGR members mainly work as adaptor proteins via the protein-protein interaction in signal transduction processes [27], since SH3 domain in a protein can easily dock to the proline-rich region in another protein. Thus, they can integrate into signaling networks anywhere in cells, depending on the cellular contexts. Here, we summarized the roles of SH3BGR members in signaling transduction processes (Figure 3). Indeed, in a study of metabolism coupling post-mitotic outgrowth to visual and sound perception network, SH3BGR was manifested as a downstream target of STAT2 [14]. We also presented that murine SH3BGRL can specifically bind to the inactive c-Src phosphorylated at tyrosine 527 via its SH3 domain to subsequently activate the downstream AKT and MAPK signaling pathways for tumorigenesis and metastasis, and breakdown of the SH3 domain in turn impeded its binding to c-Src and blocked its oncogenic role [23]. Additionally, SH3 domain is critical for SH3BP-1(SH3 binding protein 1) function in small GTPase activation, including Rac1 [28]. In line with this, forced overexpression of murine SH3BGRL in both CHO and CT-26 cells remarkably increases Rac1-GTP and cdc42-GTP activities [23]. Previously, SH3BGRL was reported to be suppressed in v-Rel mediated transformation and rescued SH3BGRL could reversely block v-Rel mediated cell transformation and tumor formation through the inhibition NF-kB signaling pathway [11], but the molecular mechanism is lacking.
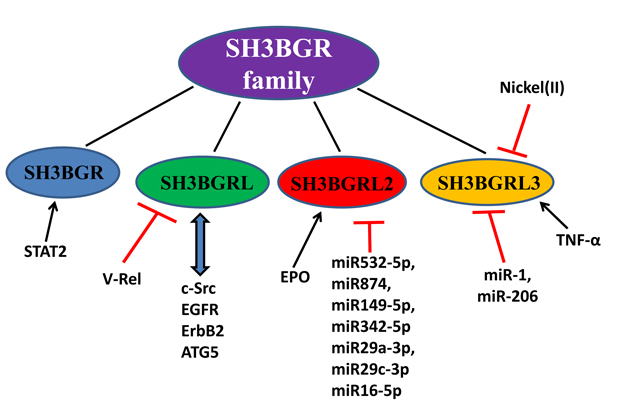
Figure 3. Signaling pathways involved with SH3BGR family members.
This figure summarizes the molecules that regulate SH3BGR family members and the signaling pathways involved with SH3BGR members, based on the published data (see Ref.). Red lines with dashes present the suppressive function; Arrows in black indicate the positive regulation, and arrows in blue stand for protein-protein interactions.
When searching for the MicroRNAs influenced by interferon-beta treatment in the peripheral blood of multiple sclerosis patients, SH3BGRL2 was found to be the target of a cluster of MicroRNAs (miR532-5p, miR874, miR149-5p, miR342-5p, miR29a-3p, miR29c-3p and miR16-5p) [29]. SH3BGRL3, also named TIP-B1, could protect cell from TNF-α induced apoptosis, which indicates its possible role in apoptosis regulation [18,19]. Additionally, SH3BGRL3 was reported to be mediated by miR-1 and/or miR-206, and its sustained expression resulted in increased proliferation and inhibition of C2C12 cell myogenesis [25].
Conclusions and expects
In this review, we summarized the big events on the characterization of SH3BGR family members and their expression patterns. With the accumulated evidence, we just preliminarily understood the SH3BGR family works mainly through their SH3 domain or proline-rich region, rather than the EVH1 domain or their thioredoxin function, in various signaling pathways as adaptor proteins. However, compared to other adaptor protein, such as Grb2, very little information on physiological and pathological roles of SH3BGR family is available. As docking proteins, their working manners should be indispensably dependent on the interacting partners in a specific cell type and the cell contexts, resulting in very complicated events in a particular disease involved with SH3BGR dysfunction, including carcinomas. Therefore, it is urgently to dissect and compare the normal and the aberrant functions of SH3BGR members in both normal and diseased tissues and cells, including their protein modification, location transportation and expression regulation, which would shed lights on the diagnosis and therapeutical reference to a related disease.
Acknowledgements
We thank Wang’s Lab members for the valuable comments and suggestions. This work was supported by Project of International Collaboration in Science and Technology of Guangdong Province (No.2014A050503030) and National Natural Science Foundation of China (No. 81171947) to WH.
References
- Pawson T, Scott JD (1997) Signaling through scaffold, anchoring, and adaptor proteins. Science 278: 2075-2080. [Crossref]
- Anderson D, Koch CA2021 Copyright OAT. All rights reserv1990) Binding of SH2 domains of phospholipase C gamma 1, GAP, and Src to activated growth factor receptors. Science 250: 979-982. [Crossref]
- Cesareni G, Panni S, Nardelli G, Castagnoli L (2002) Can we infer peptide recognition specificity mediated by SH3 domains? FEBS Lett 513: 38-44. [Crossref]
- Scartezzini P, Egeo A, Colella S, Fumagalli P, Arrigo P, et al. (1997) Cloning a new human gene from chromosome 21q22.3 encoding a glutamic acid-rich protein expressed in heart and skeletal muscle. Hum Genet 99: 387-392. [Crossref]
- Egeo A, Mazzocco M, Arrigo P, Vidal-Taboada JM, Oliva R, et al. (1998) Identification and characterization of a new human gene encoding a small protein with high homology to the proline-rich region of the SH3BGR gene. Biochem Biophys Res Commun 247: 302-306. [Crossref]
- Mazzocco M, Maffei M, Egeo A, Vergano A, Arrigo P, et al. (2002) The identification of a novel human homologue of the SH3 binding glutamic acid-rich (SH3BGR) gene establishes a new family of highly conserved small proteins related to Thioredoxin Superfamily. Gene 291: 233-239. [Crossref]
- Mazzocco M, Arrigo P, Egeo A, Maffei M, Vergano A, et al. (2001) A novel human homologue of the SH3BGR gene encodes a small protein similar to Glutaredoxin 1 of Escherichia coli. Biochem Biophys Res Commun 285: 540-545. [Crossref]
- Niebuhr K, Ebel F, Frank R, Reinhard M, Domann E, et al. (1997) A novel proline-rich motif present in ActA of Listeria monocytogenes and cytoskeletal proteins is the ligand for the EVH1 domain, a protein module present in the Ena/VASP family. EMBO J 16: 5433-5444. [Crossref]
- Holmgren A (1989) Thioredoxin and glutaredoxin systems. J Biol Chem 264: 13963-13966. [Crossref]
- Jang DG, Sim HJ, Song EK, Medina-Ruiz S, Seo JK, et al. (2015) A thioredoxin fold protein Sh3bgr regulates Enah and is necessary for proper sarcomere formation. Dev Biol 405: 1-9. [Crossref]
- Majid SM, Liss AS, You M, Bose HR (2006) The suppression of SH3BGRL is important for v-Rel-mediated transformation. Oncogene 25: 756-768. [Crossref]
- Egeo A, Di Lisi1 R, Sandri C, Mazzocco M, Lapide M, et al. (2000) Developmental expression of the SH3BGR gene, mapping to the Down syndrome heart critical region. Mech Dev 90: 313-316. [Crossref]
- Sandri C, Di Lisi R, Picard A, Argentini C, Calabria E, et al. (2004) Heart morphogenesis is not affected by overexpression of the Sh3bgr gene mapping to the Down syndrome heart critical region. Hum Genet 114: 517-519. [Crossref]
- Wang L, Huang J, Jiang M, Lin H (2012) Signal transducer and activator of transcription 2 (STAT2) metabolism coupling postmitotic outgrowth to visual and sound perception network in human left cerebrum by biocomputation. J Mol Neurosci 47: 649-658. [Crossref]
- Werner CJ, Heyny-von Haussen R, Mall G, Wolf S (2008) Proteome analysis of human substantia nigra in Parkinson's disease. Proteome Sci 6: 8. [Crossref]
- De Andrade T, Moreira L, Duarte A, Lanaro C, De Albuquerque D, et al. (2010) Expression of new red cell-related genes in erythroid differentiation. Biochem Genet 48: 164-171. [Crossref]
- Collares CV, Evangelista AF, Xavier DJ, Takahashi P, Almeida R, et al. (2013) Transcriptome meta-analysis of peripheral lymphomononuclear cells indicates that gestational diabetes is closer to type 1 diabetes than to type 2 diabetes mellitus. Mol Biol Rep 40: 5351-5358. [Crossref]
- Berleth ES, Nadadur S, Henn AD, Eppolito C, Shiojiri S, et al. (1999) Identification, characterization, and cloning of TIP-B1, a novel protein inhibitor of tumor necrosis factor-induced lysis. Cancer Res 59: 5497-5506. [Crossref]
- Berleth ES, Henn AD, Gurtoo HL, Wollman R, Alderfer JL, et al. (2000) A novel tumor necrosis factor-alpha inhibitory protein, TIP-B1. Int J Immunopharmacol 22: 1137-1142. [Crossref]
- Deng YJ, Huang ZX, Zhou CJ, Wang JW, You Y, et al. (2006) Gene profiling involved in immature CD4+ T lymphocyte responsible for systemic lupus erythematosus. Mol Immunol 43: 1497-1507. [Crossref]
- van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, et al. (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415: 530-536. [Crossref]
- Abba MC, Hu Y, Sun H, Drake JA, Gaddis S, et al. (2005) Gene expression signature of estrogen receptor alpha status in breast cancer. BMC Genomics 6: 37. [Crossref]
- Wang H, Liu B, Al-Aidaroos AQ, Shi H, Li L, et al. (2015) Dual-faced SH3BGRL: oncogenic in mice, tumor suppressive in humans. Oncogene. [Crossref]
- Lee SH (2006) Differential gene expression in nickel(II)-treated normal rat kidney cells. Res Commun Mol Pathol Pharmacol 119: 77-87. [Crossref]
- Goljanek-Whysall K, Pais H, Rathjen T, Sweetman D, Dalmay T, et al. (2012) Regulation of multiple target genes by miR-1 and miR-206 is pivotal for C2C12 myoblast differentiation. J Cell Sci 125: 3590-3600. [Crossref]
- Chiang CY, Pan CC, Chang HY, Lai MD, Tzai TS, et al. (2015) SH3BGRL3 Protein as a Potential Prognostic Biomarker for Urothelial Carcinoma: A Novel Binding Partner of Epidermal Growth Factor Receptor. Clin Cancer Res 21: 5601-5611. [Crossref]
- Mayer BJ, Baltimore D (1993) Signalling through SH2 and SH3 domains. Trends Cell Biol 3: 8-13. [Crossref]
- Cicchetti P, Mayer BJ, Thiel G, Baltimore D (1992) Identification of a protein that binds to the SH3 region of Abl and is similar to Bcr and GAP-rho. Science 257: 803-806. [Crossref]
- Hecker M, Thamilarasan M, Koczan D, Schroder I, Flechtner K, et al. (2013) MicroRNA expression changes during interferon-beta treatment in the peripheral blood of multiple sclerosis patients. Int J Mol Sci 14: 16087-16110. [Crossref]



