MicroRNAs (miRNAs) are an extensive class of small noncoding RNAs, having profound impact on many biological processes in development, differentiation, growth and metabolism by regulating gene expression. Polymorphism in 3’UTR showed to alter the miRNA binding site in several human genes and consequently affects the function of the resulting protein posttrancriptionally. To investigate the putative functional consequences of the Cyclin D1 (CCND1) G/C1722 SNP (rs678653) located on 3’UTR of the gene, in-silico approach was applied for the prediction of miRNA binding sites using miRDB version 3.0 & 5.0.
We were able to predict 16 miRNA/miRNA target sites on 3’UTR of human CCND1 gene. Out of 16 predicted miRNAs 11 were novel miRNAs/miRNA target sites which target CCND1. However no miRNA target sites were predicted on the DNA/RNA sequence of 3’UTR of human CCND1 gene encompassing the SNP (rs678653). Hence, SNP (rs678653) is not likely to be of functional importance according to this algorithmic prediction. Further experimental analysis is required to validate this computational prediction for CCND1 variants.
miRNA, SNP, CCND1, miRDB, microsniper, rs678653, 3’UTR, miRNA target sites, microRNA, cyclin D1
Abbreviations
miRNA: MicroRNA; SNP: Single nucleotide polymorphism; CCND1: Cyclin D1, UTR: Untranslated region; SVM: Support vector machine
MicroRNAs (miRNAs) have emerged as an important class of short endogenous non-coding RNAs, 18-25 nucleotide (nt) that act as post-transcriptional regulators of gene expression. miRNAs are thought to be involved in diverse biological processes by regulating gene expression. Numerous miRNAs have been identified in various species and many more miRNAs remain to be detected [1].
Following the first documentation of miRNA in C. elegans about two decades ago by Lee et al. [2] to date more than 4000 distinct miRNAs have been annotated in vertebrates, invertebrates and plants, and many miRNAs that correspond to putative genes have also been identified. This discovery has introduced a totally new dimension in our understanding of how gene expression is regulated [3].
In the cytoplasm of the cell, miRNA is typically found in the mature form as 18-25 nt long. miRNAs are initially transcribed as part of much longer primary transcripts (termed pri-miRNAs) [4]. Pri-miRNA is ~100 bp in length and forms a stem-loop fold back structure. It is further processed into a mature miRNA (MIR) that is ~22 bp in size by RNAse III ribonucleases and binds to a specific target site on an mRNA for posttranscriptional repression. The critical region for MIR binding in animals is the “seed” region (nt 2-7 from 5’ end of MIR), which preferentially binds to a target site in the 3’UTR of the mRNA by Watson-Crick pairing [5].
SNPs associated with polygenetic disorders such as cancer can create, destroy or modify miRNA binding sites. Genome-wide bioinformatics analysis predicted ∼64% of transcribed SNPs as target SNPs that can modify (increase/decrease) the binding energy of putative miRNA:: mRNA duplexes by >90% [6].
Previously several reports have shown that miRNA signatures are emerging as a potential biomarker. Numerous miRNAs have been found to have links with some types of cancer [7,8]. These types of studies were mainly remained focussed on cancers including papillary thyroid and colorectal carcinoma [7,9,10], thereby showing the importance of exploring the function of miRNAs in the field of oncology.
Various efforts have been made to evaluate the effect of SNPs on putative miRNA target sites on cancer susceptibility e.g. breast cancer [6,11] non-small cell lung cancer [12], esophageal squamous cell carcinoma [13], gastric cancer [14], colon cancer [15], head and neck cancer [16], glioma [17], hepatocellular carcinoma [18], thyroid cancer [19], prostate cancer [20], renal cell carcinoma [21] and bladder cancer [22]. A case-control population study has been conducted by Nicoloso et al. [6], for assessment of implication of target SNPs on breast cancer susceptibility and observed that germline occurrence of rs799917-BRCA1 and rs334348-TGFR1 significantly varies among populations with different risks of developing breast cancer.
There are several databases available for the prediction of miRNA target sites including miRDB. It was created by Xiaowei Wang's lab at the Department of Radiation Oncology, Washington University School of Medicine in St. Louis [23,24]. It is an online database for the prediction of miRNA target sites and functional annotations in animals. All the targets were predicted by a new bioinformatics tool, which was developed by analyzing thousands of genes impacted by miRNAs with a Support Vector Machine (SVM). Common features associated with miRNA target binding have been identified and used to predict miRNA targets. miRDB hosts predicted miRNA targets for five species including human, mouse, rat, dog and chicken.
Cyclin D1 belongs to the core cell cycle machinery, and it is frequently overexpressed in human cancers. The full repertoire of cyclin D1 functions in normal development and oncogenesis is unclear at present [25].
Previously we have reported a potential protective effect of the CCND1 SNP G/C1722 (rs678653) on 3’UTR in cervical cancer susceptibility in Indian population [26]. Jiang et al. [27] recently, published a report concerning the role of miRNAs which target CCND1, but they have not included the association of any CCND1 SNP in their study.
To the best of our knowledge, there is no report available addressing the functional importance of CCND1 gene polymorphism rs678653 located on 3’UTR.
It is well established fact that when SNPs occur in 3’UTRs they interfere with mRNA stability and translation by altering polyadenylation, protein:mRNA and miRNA::mRNA regulatory interactions [6]. It has also been revealed that the 3’ UTRs of human protein-coding genes (for example CCND1) are rich in miRNA target sites. So, with this background we hypothesised that G-C nucleotide exchange at 3’UTR of CCND1 generated by SNP rs678653 could influence the putative 3’ binding site of the miRNAs.
Prediction of novel miRNA/miRNA target sites
We were able to predict 16 miRNA target sites on 3’UTR of human CCND1 gene with the help of miRDB version 3.0. Among 16 miRNAs, eleven (miR-1237, miR-548e, miR-522, miR-942, miR-548g, miR-891b, miR-1283, miR-495 and miR-1281, miR-147, miR-584f ) were novel which we have discovered first time in the present study, while 5 miRNAs (miR-374, miR-548d, miR-219-1-3p, miR-340 and miR-511) were found to be overlapped with TargetScan and miRanda database. Based on release of the database on April 2009 for putative binding of 3’UTR of CCND1 gene, hsa-miR-147 had the highest score (87), of 16 predicted miRNAs with calculated scores in the range of 87-53 (Table 1).
Table 1. CCND1 (Gene 595) is predicted to be targeted by 16 miRNAs in miRDB
Target Detail (seed location) |
Target Rank |
Target Score |
miRNA Name |
miRNA Seq1 |
1617, 2294 |
1 |
87 |
hsa-miR-147 |
GUGUGUGGAAAUGCUUCUGC |
1338, 1375 |
2 |
84 |
hsa-miR-548d-3p |
CAAAAACCACAGUUUCUUUUGC |
100 |
3 |
79 |
hsa-miR-1237 |
UCCUUCUGCUCCGUCCCCCAG |
309 |
4 |
78 |
hsa-miR-219-1-3p |
AGAGUUGAGUCUGGACGUCCCG |
1623, 3149 |
5 |
73 |
hsa-miR-511 |
GUGUCUUUUGCUCUGCAGUCA |
1074, 2050 |
6 |
71 |
hsa-miR-548e |
AAAAACUGAGACUACUUUUGCA |
460 |
7 |
71 |
hsa-miR-522 |
AAAAUGGUUCCCUUUAGAGUGU |
833 |
8 |
69 |
hsa-miR-942 |
UCUUCUCUGUUUUGGCCAUGUG |
1554 |
9 |
67 |
hsa-miR-548g |
AAAACUGUAAUUACUUUUGUAC |
1074, 2050 |
10 |
64 |
hsa-miR-548f |
AAAAACUGUAAUUACUUUU |
2477 |
11 |
63 |
hsa-miR-891b |
UGCAACUUACCUGAGUCAUUGA |
885, 2131 |
12 |
58 |
hsa-miR-1283 |
UCUACAAAGGAAAGCGCUUUCU |
585 |
13 |
55 |
hsa-miR-340 |
UUAUAAAGCAAUGAGACUGAUU |
605, 2501 |
14 |
54 |
hsa-miR-374a |
UUAUAAUACAACCUGAUAAGUG |
2632 |
15 |
53 |
hsa-miR-495 |
AAACAAACAUGGUGCACUUCUU |
370 |
16 |
50 |
hsa-miR-1281 |
UCGCCUCCUCCUCUCCC |
Note: 1miRNA seed region: 7 nucleotides long, accession No. NM_053056
By using miRDB v.5 released on August 2014, we found that CCND1 gene is predicted to be targeted by 115 miRNAs (data not shown).
No miRNA target sites were predicted on the DNA/RNA sequence encompassing the G/C1722 SNP (rs678653) by both the versions of the database. Two miRNAs (hsa-miR-374a and hsa-miR-942) were found to be resides on flanking sequences of the SNP rs678653 site.
By selecting the details option in the output window the results displayed for respective miRNAs. Here we have presented information only for hsa-miR-147 which scored highest (87) in the target list, (Supplementary Data 1). Similarly, the details for each predicted miRNA was obtained and their respective seed regions (7 nt long) were located on the 3’UTR sequence of CCND1 (Supplementary Data 2).
Prediction of functional effect of CCND1 SNP (rs678653)
Computational prediction based on MicroSNiPer database showed lack of functional effect of CCND1 polymorphism located on 3’UTR (rs678653). This observation is consistent with miRDB findings and could further be strengthen from the algorithmic results which showed the absence of any putative miRNA/miRNA target site on the nucleotide sequence encircling the polymorphic site on CCND1.
After the discovery of miRNAs about two decades ago their potential role has been shown in different diseases and hence added further impetus in miRNA research [2]. miRNAs function as an important posttranscriptional regulators of mRNA expression by binding to the 3’UTR and repressing translation [28,29]. A SNP that alters the true target site may have biological implications [5]. Since the 3’UTR is evolving as a hotspot for pathology, therefore, SNPs at miRNA target sites may be helpful in distinguishing various cancers and diseases [30].
At present various databases are freely available for the prediction of miRNA target site. Most of them are based as online search while few need to be downloaded in order to access.
We chose miRDB system for prediction of miRNA target sites with reference to CCND1, because this is the one of the latest database freely available as an online search and their design strategy is based on mature miRNAs unlikely most of the other platforms which relies on pre-miRNAs. Because it is the mature miRNA, not their precursors, which are directly involved in the target down-regulation [23].
On comparison of the miRDB output from other popular databases such as PicTar, miRanda, TargetScan, only two miRNAs (miR-374 and miR-548) were commonly present among miRanda and TargetScan. While three miRNAs (miR-219-1-3p, miR-340 and miR-511) were found to be overlapped in miRanda and miRDB programme. However it was quite surprising to note that not a single miRNA was found to be predicted commonly between miRDB and PicTar (Table 2, Figure 1).
Table 2. Comparison of CCND1 miRNAs predicted from different databases
Name of the Database |
Name of the miRNAs |
|
miR-374 |
miR-548d |
miR-219-1-3p |
miR-340 |
miR-511 |
miRDB1 |
ü |
ü |
ü |
ü |
ü |
miRanda2 |
X |
X |
ü |
ü |
ü |
TargetScan3 |
ü |
ü |
X |
X |
X |
List of novel miRNAs predicted by miRDB In the present study |
miR-147 miR-1237 miR-548e miR-522 miR-942 miR-548g |
miR-548f miR-891b miR-1283 miR-495 miR-1281 |
Note: 1-Present study (http://mirdb.org), 2,3-Jiang et al., 2009, (http://www.targetscan.org) (http://microrna.org)
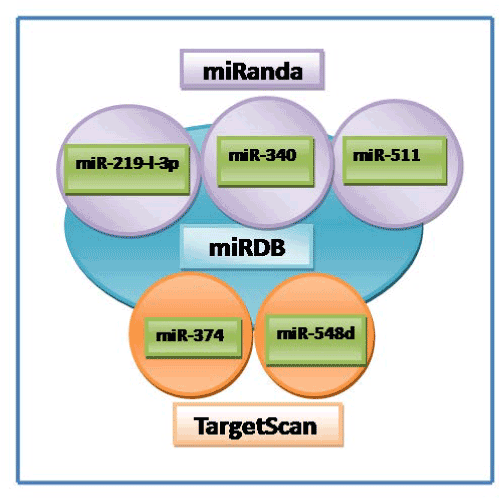
Figure 1. Overlap of CCND1 miRNAs predicted from three different databases (miRDB/miRanda/TargetScan)
However, few studies have thrown some light on the prediction of target sites on CCND1 gene. The first systematic approach for validation of the predicted miRNAs for cyclin D1 was carried out by Jiang et al. [27]. By using a reporter system, this research group showed that only a small fraction of predicted miRNA/mRNA interactions are experimentally valid in case of CCND1.
Till date, no effort has been made to study the influence of CCND1 3’UTR SNP in this context. To the best of our knowledge, this is the first attempt to apply miRDB v.3/v.5 database [31] to predict the miRNA target sites on CCND1 in correlation with SNP (rs678653) located on 3’UTR of the gene. It was mentioned in one study that polymorphisms in predicted miRNA binding sites are likely to be deleterious; they are candidates for causal variants of human disease [32]. In our previous study, we showed that CCND1 gene SNP (rs678653) might provide some protection against the development of cervical cancer in Indian population [26].
Our computational analysis revealed that SNP (rs678653) located on 3’UTR of CCND1 is not presenting the target site for any of the 16 predicted miRNAs listed in Table 1. However, no solid conclusion can be drawn just on the basis of computational algorithm. Therefore, to confirm this bioinformatics’ data, further experimental validation is warranted to investigate the factual functional relevance of CCND1 SNP (rs678653) posttranscriptionally.
Taken together, this pooled prediction based on algorithmic computational evidence of the CCND1 variant suggesting the possible explanation that SNP (rs678653) on 3’UTR of the gene is less likely to be of functional importance. This may be one of the reasons for having limited reports available addressing the role of CCND1 SNP (rs678653) in comparison to SNP (rs9344) G870A, which is widely studied in various cancers and other diseases because of the functional implications.
In this study, we have identified 11 novel miRNAs/miRNA target sites by miRDB, but no miRNA binding sites were found to be located on the DNA/RNA sequence of 3’UTR of human CCND1 gene encompassing the SNP (rs678653). Therefore, this polymorphism does not seem to have any functional effect, but still, further experimental analysis is required to validate this computational prediction for CCND1 variants, as it is evident from the literature that the ratio of predicted and validated miRNA target sites is very low.
Prediction of miRNA target sites using miRDB
To investigate the putative functional consequences of the human CCND1 SNP rs678653 located on 3’UTR of the gene, in-silico analysis (Bioinformatic approach) was applied for the prediction of miRNA binding sites using miRDB version 3.0 released on April 2009 (http://mirdb.org), last updated on 26 August 2010. Schematic representation of methodology used in the present study is shown in Figure 2.
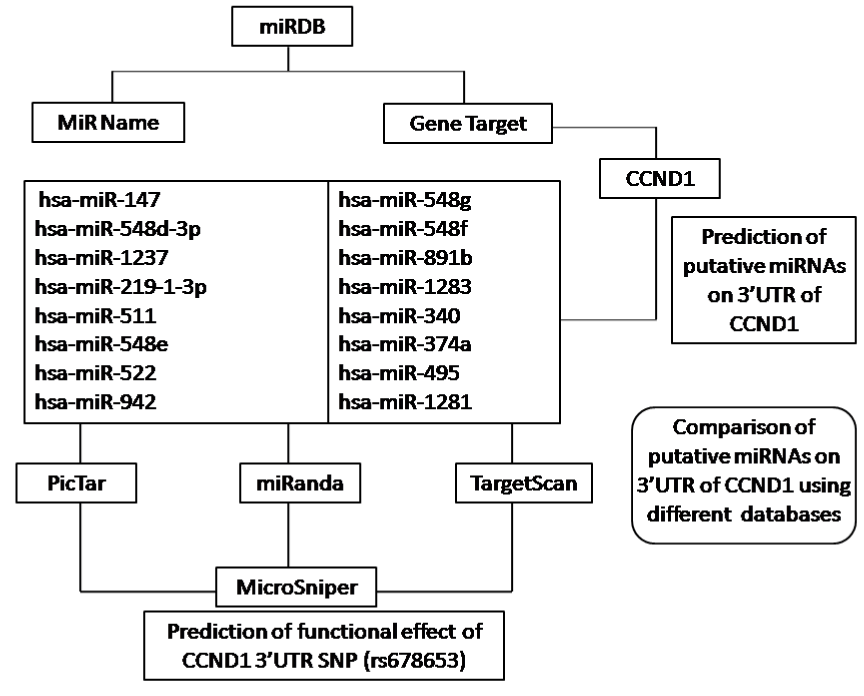
Figure 2. Flow diagram of methodology
The miRDB utilizes the MirTarget 2 prediction tool and miRBase 13.0 as a miRNA source. miRDB version 3.0 contains 2,295 miRNAs targeting 58,953 unique genes for five species including human, mouse, rat, dog and chicken. Specifically, for Homo sapiens it is having 703 miRNAs targeting 16,856 unique genes. All the predicted miRNAs and their target sites were searched via freely accessible online database. miRDB v 3.0 displays a web query interface with the two alternatives- first, miRNA name and second, gene target. We used the gene target (i.e., CCND1) as a web query, and the result was displayed in the descending order of the predicted target scores.
We have also performed this analysis by latest miRDB version 5.0 last modified on March 10, 2015.
Interpretation of target scores
All the predicted targets have target prediction scores between 50 and 100. These scores are assigned by the new computational target prediction algorithm. The higher the score, the more confidence we have in this prediction. That is why the search result is ordered by prediction score. A predicted target with prediction score >80 is most likely to be real. If the score is <60, one should be cautious and it is recommended to have other supporting evidence as well.
Comparison of putative CCND1 miRNAs predicted by different databases
The miRDB output was compared with other popular databases such as PicTar (http://www.pictar.org), miRanda (http://microrna.org) and TargetScan (http://www.targetscan.org) (Table 2, Figure 1).
Prediction of functional effect of CCND1 SNP (rs678653)
2021 Copyright OAT. All rights reserv
For the prediction of functional effect of CCND1 SNP (rs678653) on 16 identified putative microRNA targets on the 3’UTR of the gene, a computational analysis was performed by using MicroSNiPer. It is a web based tool for the prediction of functional implication of single nucleotide variation on putative miRNA target sites which was recently introduced by Barenboim et al. [33]. This database interrogates the 3'-untranslated region and predicts whether a SNP within the target site will disrupt/eliminate or enhance/create a microRNA binding site. MicroSNiPer computes these sites and explores the effects of SNPs in real time. MicroSNiPer is freely accessible at http://cbdb.nimh.nih.gov/microsniper.
The authors acknowledge miRDB (http://mirdb.org) and MicroSNiPer (http://cbdb.nimh.nih.gov/microsniper) which was accessed for the predictions of novel miRNAs/miRNA target sites and functional effect of the SNP. . This study was supported by NICPR (ICMR) Noida.
The full length 3’UTR sequence showing the miRNAs/miRNA target sites of CCND1 as a supplementary data is available online. Availability: miRDB v.3/v.5 database is freely accessible at (http://mirdb.org). Supplementary Data (1): Representative Description of Predicted MicroRNA and Target Gene
Supplimentary Data (2): 3' UTR Sequence Showing the Predicted Target Sites for all the 16 miRNAs of CCND1.
No competing financial interest reported.
View supplementary data
- Liu Q, Fu H, Sun F, Zhang H, Tie Y, et al. (2008) miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res 36: 5391-5404. [Crossref]
- Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843-854. [Crossref]
- Sturm M, Hackenberg M, Langenberger D, Frishman D (2010) TargetSpy: a supervised machine learning approach for microRNA target prediction. BMC Bioinformatics 11: 292. [Crossref]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN (2002) MicroRNA maturation: stepwise processing and subcellular localization. EMBOJ 21: 4663-4670. [Crossref]
- Saunders MA, Liang H, Li WH (2007) Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci U S A 104: 3300-3305. [Crossref]
- Nicoloso MS, Sun H, Spizzo R, Kim H, Wickramasinghe P, et al. (2010) Single-nucleotide polymorphisms inside MicroRNA target sites influence tumor susceptibility. Cancer Res 70: 2789-2798. [Crossref]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu S, et al. (2005) A microRNA polycistron as a potential human oncogene. Nature 435: 828–833. [Crossref]
- Mraz M, Pospisilova S, Malinova K, Slapak I, Mayer J, (2009) MicroRNAs in lymphocytic leukemia pathogenesis and disease subtypes. Leuk Lymphoma 50: 506–509.
- Croce CM, Calin GA (2005) miRNAs, cancer, and stem cell division. Cell 122: 6-7. [Crossref]
- Cummins JM, Velculescu VE (2006) Implications of micro-RNA profiling for cancer diagnosis. Oncogene 25: 6220-6227. [Crossref]
- Gao LB, Bai P, Pan XM, Jia J, Li LJ, et al. (2011) The association between two polymorphisms in pre-miRNAs and breast cancer risk: a meta-analysis. Breast Cancer Res Treat 125: 571-574. [Crossref]
- Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, et al. (2008) A SNP in a let-7 microRNA complementary site in the KRAS 3' untranslated region increases non-small cell lung cancer risk. Cancer Res 68: 8535-8540. Crossref
- Guo H, Wang K, Xiong G, Hu H, Wang D, et al. (2010) A functional varient in microRNA-146a is associated with risk of oesophageal squamous cell carcinoma in Chinese Han. Fam Cancer 9: 599-603. [Crossref]
- Sun Q, Gu H, Zeng Y, Xia Y, Wang Y, et al. (2010) Hsa-mir-27a genetic variant contributes to gastric cancer susceptibility through affecting miR-27a and target gene expression. Cancer Sci 101: 2241-2247. [Corssref]
- Boni V, Zarate R, Villa JC, Bandrés E, Gomez MA, et al. (2011) Role of primary miRNA polymorphic variants in metastatic colon cancer patients treated with 5-fluorouracil and irinotecan. Pharmacogenomics J 11: 429-436. [Crossref]
- Liu Z, Li G, Wei S, Niu J, El-Naggar AK, et al. (2010) Genetic variants in selected pre-microRNA genes and the risk of squamous cell carcinoma of the head and neck. Cancer 116: 4753-4760. [Crossref]
- Dou T, Wu Q, Chen X, Ribas J, Ni X, et al. (2010) A polymorphism of microRNA196a genome region was associated with decreased risk of glioma in Chinese population. J Cancer Res Clin Oncol 136: 1853-1859. [Crossref]
- Qi P, Dou TH, Geng L, Zhou FG, Gu X, et al. (2010) Association of a variant in MIR 196A2 with susceptibility to hepatocellular carcinoma in male Chinese patients with chronic hepatitis B virus infection. Hum Immunol 71(6): 621-626. [Crossref]
- Jazdzewski K, Liyanarachchi S, Swierniak M, Pachucki J, Ringel MD, et al. (2009) Polymorphic mature microRNAs from passenger strand of pre-miR-146a contribute to thyroid cancer. Proc Natl Acad Sci U S A 106: 1502-1505. [Crossref]
- Sun J, Zheng SL, Wiklund F, Isaacs SD, Li G, et al. (2009) Sequence variants at 22q13 are associated with prostate cancer risk. Cancer Res 69: 10-15. [Crossref]
- Horikawa Y, Wood CG, Yang H, Zhao H, Ye Y, et al. (2008) Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin Cancer Res 14(23):7956-7962. [Crossref]
- Yang H, Dinney CP, Ye Y, Zhu Y, Grossman HB, et al. (2008) Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer. Cancer Res 68: 2530-2537. [Crossref]
- Wang X, El Naqa IM (2008) Prediction of both conserved and non-conserved microRNA targets in animals. Bioinformatics 24: 325-332. [Crossref]
- Wang X (2008) miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA 14: 1012-1017. [Crossref]
- Bienvenu F, Jirawatnotai S, Elias JE, Meyer CA, Mizeracka K, et al. (2010) Transcriptional role of cyclin D1 in development revealed by a genetic–proteomic screen. Nature 463: 374-378.
- Thakur N, Hussain S, Kohaar I, Tabassum R, Nasare V, et al. (2009) Genetic variant of CCND1 association with HPV-mediated cervical cancer in Indian population. Biomarkers 14(4): 219-225. [Crossref]
- Jiang Q, Feng MG, Mo YY (2009) Systematic validation of predicted microRNAs for cyclin D1. BMC Cancer 9: 194. [Crossref]
- Ambros V (2004) The functions of animal microRNAs. Nature 431: 350-355. [Crossref]
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281-297. [Crossref]
- Mishra PJ, Mishra PJ, Banerjee D, Bertino JR (2008) miRSNPs or miR-polymorphisms, new players in microRNA mediated regulation of the cell: Introducing microRNA pharmacogenomics. Cell Cycle 7(7): 853-858. [Crossref]
- Wong N, Wang X (2015) miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 43: D146-152. [Crossref]
- Chen K, Rajewsky N (2006) Natural selection on human microRNA binding sites inferred from SNP data. Nature Genetics 38(12): 1452-1456. [Crossref]
- Barenboim M, Zoltick BJ, Guo Y, Weinberger DR (2010) MicroSNiPer: a web tool for prediction of SNP effects on putative microRNA targets. Hum Mutat 31: 1223-1232. [Crossref]


