The need for a Next-Generation Sequencing workflow is rapidly increasing as the sequencing technology matures and becomes more feasible as a diagnostic tool. Characterizing the tumor profile with as much information as possible is important to identify patient populations that may be poor responders to standard therapies or would benefit from alternative therapeutic strategies. Driven by tissue context, sequencing joined with immuno-histochemical tests for specific markers can provide comprehensive information to inform clinicians on potential treatment pathways. We have developed an integrated workflow solution that combines the use of IHC, digital imaging, automated dissection of FFPE and NGS to capture important information to drive therapeutic opportunities. To evaluate the workflow, we sequenced both isolated tumor regions and whole tissue resections from the same case. We were able to show that samples acquired from automated dissection are suitable for use in an NGS workflow and can provide comprehensive information about the tumor profile and additional diagnostic IHC markers. Additionally, we found that the use of automated dissection of tumor ROIs can allow for detection of variants that would otherwise go undetected in 70% of the cases, several of which were clinically actionable. Collectively, these results suggest that Integrated CDx is a powerful approach to characterizing a specific tumor sample with the potential to identify effective cancer therapies in the realm of personalized healthcare.
NGS, dissection, companion diagnostics
Abbreviations: CDx: Companion Diagnostics, DAB: Diaminobenzidine, DNA: Deoxyribonucleic Acid, FFPE: Formalin Fixed Paraffin Embedded, FOV: Frequency of Variation, IHC: Immunohistochemistry, NGS: Next-Generation Sequencing, NSCLC: Non Small Cell Lung Cancer, ROI: Region of Interest
Immunohistochemistry (IHC) of single biomarkers has proven to be an effective mechanism to identify patients as candidates for targeted therapy. However, this method alone has limitations in scope and application. A single IHC stain renders a snapshot of biomarker expression in a tumor and does not describe the complete profile. Often, patients’ tumor samples are limited and tumor heterogeneity is extremely common [1-3]. Intra-tumor heterogeneity is the presence of more than one clonal subtype of cancer within a tumor and has been found to be common in multiple tumor types [4-6]. This type of heterogeneity can affect response to treatment, including resistance to drugs or only partial response [5,6]. The recent development of deep sequencing has supported this idea by highlighting genetic heterogeneity [7,8]. However, with sequencing alone, there can be a loss of tissue context. Precision dissection of tumors has been successfully applied to deal with tumor heterogeneity [9].
These factors suggest an integrated diagnostic would be an attractive approach to interrogate biological drivers of tumor growth, while preserving the sample to evaluate the candidacy of the patient for additional personalized investigational therapies. This integrated diagnostic pathway can be accomplished by combining molecular characterization and IHC stains to determine the best treatment options.
In this “proof-of-principle” study, we present an evaluation of the molecular profile of multiple tumor cases that showed intra-tumor heterogeneity based on a single IHC stain. We successfully performed targeted sequencing on specific regions of interest (ROI) within the tumor. In some cases, additional IHC stains were performed based on analysis of the sequencing data. Altogether, we were able to confirm that molecular data combined with IHC data could be used to fully characterize tumor specimens and that additional data could affect treatment options.
Sample acquisition
Human formalin-fixed paraffin embedded (FFPE) tissue blocks (Non-small cell lung cancer, prostate cancer, and head & neck cancer) were acquired from internal sources. Blocks were de-identified and cannot be linked to treatment or survival.
Tissue staining and automated dissection
All immuno-histochemistry was performed on the BenchMark Ultra automated staining platform from Ventana Medical Systems with the OptiView Universal DAB Detection Kit (VMSI, Catalog No. 760-500). Prior to staining, 4 µm sections were mounted on superfrost plus slides. The following antibodies from Ventana Medical Systems were used according to package insert conditions: anti-EGFR (5B7) Rabbit Monoclonal Primary Antibody (790-4347), anti-EGFR L858R (SP125) Rabbit Monoclonal Primary Antibody (790-4649), anti-HER-2/neu (4B5) Rabbit Monoclonal Primary Antibody (790-2991), and anti-p53 (Bp53-11) Primary Antibody (760-2542). All IHC staining protocols have been validated for use on clinical specimens in Ventana Medical System’s CAP/CLIA laboratory. Tumor ROIs were annotated by a qualified reader. Prior to automated dissection, 4 µm sections were mounted on slides and de-paraffinized. Dissection was performed on a Roche Automated Dissection platform, and ROIs were collected into a Tris-based buffer. Tissue from whole resections was collected from two 20 µm scrolls cut off the FFPE block with a microtome.
DNA extraction and quantification
Extraction of DNA from captured FFPE was completed with the Roche MagNA Pure 96 Instrument using the DNA Tissue FFPE SV 2.0 protocol and the MagNA Pure 96 DNA and Viral NA Small Volume Kit (Roche, Catalog No. 06543588001). Quantification of amplifiable DNA was performed via qPCR on the Roche Lightcyler 480 and Kapa Biosystem’s hgDNA Quantification and QC Kit (Kapa Biosystems, Catalog No. KK4963).
Library construction and sequencing
The AmpliSeq/PGM sequencing technology was used for comparison of tumor profiles. DNA from captured tumor or the whole specimen, was constructed using the Ion AmpliSeqTM Cancer Hotspot Panel v2 from Thermo Fisher. The panel contains 207 primer pairs and allows for coverage of hotspots in 50 genes (ABL1, AKT1, ALK, APC, ATM, BRAF, CDH1, CDKN2A, CSF1R, CTNNB1, EGFR, ERBB2, ERBB4, EZH2, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, GNA11, GNAS, GNAQ, HNF1A, HRAS, IDH1, JAK2, JAK3, IDH2, KDR, KIT, KRAS, MET, MLH1, MPL, NOTCH1, NMP1, NRAS, PDGFRA, PIK3CA, PTEN, PTPN11, RB1, RET, SMAD4, SMARCB1, SMO, SRC, STK11, TP53, VHL). The panel and assay conditions were optimized and validated in-house. Using 10ng of input DNA, library preparation and barcoding was carried out on Thermo Fisher’s ION Chef, utilizing the AmpliSeqTM DL8 Kit (Thermo Fisher, Catalog No. A29024) and following the manufacturer’s recommended protocol. Template preparation and chip loading was also performed on the ION Chef (software version 5.0.2), using the Hi-Q Chef Kit (Thermo Fisher, Catalog No. A25948) and a concentration of 35pM for the pooled libraries. A 318BC chip was used with a max of 8 barcoded samples. Sequencing was performed on Thermo Fisher’s Ion Torrent PGM, utilizing the Torrent Suite (software version 5.0.4) and Variant Caller Plugin for analysis. Filters utilized during analysis include ≥500X coverage and ≥1% frequencing of variation (FOV).
Integrated diagnostic workflow
Development of an integrated approach to diagnosis offers the ability to provide comprehensive information about clinical samples to aid in treatment decisions and prognosis. Our Integrated Companion Diagnostics (ICDx) workflow is a multistep approach that leverages the effectiveness and contextual advantages of IHC with the depth of analysis of next generation sequencing (NGS). Following staining of a section for the primary biomarker, selected tumor region(s) of interest can be excised on subsequent, unstained, serial sections by automated dissection. The captured FFPE tissue is put through a fully automated NGS workflow including DNA extraction, library prep, and QC. Through analysis of sequencing results, additional candidate biomarkers of interest with potential as tumor drivers and candidate proteins for targeted therapy can be identified, leading to expansion of companion diagnostic hypotheses and the development of new single marker or multiplex IHC assays.
ROIs were selected based on primary IHC marker staining of PTEN, EGFR, or PD-L1. Tumor regions negative for the primary marker (positive in the case of PTEN) were annotated for isolation. These regions were selected due to the fact that the primary marker was not a driver of that tumor region. The majority of regions isolated from FFPE via automated dissection (with adequate DNA based on qPCR measurements) were successful in downstream library prep and sequencing utilizing the Ion Torrent PGM. From the initial samples used in isolation and DNA extraction, 10/11 (90.9%) were successfully sequenced on the PGM with the Cancer Hotspot Panel. Total ROI tissue size, tumor type, and resulting DNA amount isolated from the automated MagnaPure96 can be found in Table 1. DNA yields ranged from 15ng – 337ng despite similar size regions used for isolation. This is indicative of the variation that is seen in DNA quality with FFPE tissues. The single sample that failed sequencing was a NSCLC specimen and yielded only 18ng of total DNA.
Table 1. Summary of specimen details regarding ROI and DNA measurements
| Sample |
Specimen Type |
Total ROI Isolated (mm2) |
DNA (ng) |
Primary IHC Marker |
| 1 |
Prostate |
196 |
57.0 |
PTEN |
| 2 |
Prostate |
240 |
37.5 |
PTEN |
| 3 |
Prostate |
256 |
240.0 |
PTEN |
| 4 |
Prostate |
245 |
160.0 |
PTEN |
| 5 |
NSCLC |
241 |
15.0 |
EGFR |
| 6 |
NSCLC |
239 |
18.0 |
EGFR |
| 7 |
NSCLC |
241 |
78.5 |
EGFR |
| 8 |
H&N |
223 |
188.5 |
PD-L1 |
| 9 |
H&N |
166 |
69.0 |
PD-L1 |
| 10 |
H&N |
267 |
337.5 |
PD-L1 |
| 11 |
H&N |
232 |
198.0 |
PD-L1 |
Comparison of NGS data from isolated tumor region and whole resection
Sequencing of either an isolated ROI or the whole tissue from the same specimen generally yielded very different molecular profiles. Of the specimens sequenced 8/10 (80%) had an increased number of mutations identified in the tumor ROI than were identified in the whole specimen (Table 2). There was some overlap of mutations present in both the ROI and the whole specimen, generally for SNPs present at higher FOV (data not shown). Of the mutations present in the ROIs isolated versus whole specimens, 7/10 (70%) of the cases had actionable hotspot mutations in the ROI not identified when sequencing the whole specimen. These included 3 prostate specimens, 2 NSCLC specimens, and 2 H&N specimens.
Table 2. Summary of total variants identified in ROI versus whole specimen
| Sample |
ROI SNPs |
Whole Specimen SNPs |
Overlapping SNPs (concordance %) |
ROI % of Whole Specimen |
| 1 |
20 |
8 |
7 (35%) |
21% |
| 2 |
12 |
10 |
7 (58%) |
15% |
| 3 |
19 |
14 |
5 (26%) |
17% |
| 4 |
13 |
7 |
2 (15%) |
16% |
| 5 |
25 |
8 |
8 (32%) |
11% |
| 6 |
n/a |
n/a |
n/a |
26% |
| 7 |
26 |
12 |
11 (42%) |
50% |
| 8 |
10 |
18 |
7 (39%) |
54% |
| 9 |
16 |
18 |
11 (61%) |
51% |
| 10 |
8 |
6 |
6 (75%) |
29% |
| 11 |
17 |
6 |
6(35%) |
82% |
Evaluation of secondary IHC markers
A variety of mutations were identified in the sequencing profile of the ROIs selected (Figure 1). Since the ROIs were tumor regions not driven by the initial primary IHC marker, the sequencing profiles were evaluated for additional indications of potential diagnostic markers that may be tested via a clinically validated IHC method. Abnormal expression of at least one protein was confirmed in 4 (40%) of the cases, which also had mutations in the coding gene. These genes include EGFR, ERBB2, EZH2, and TP53. Not all mutations identified in these cases resulted in confirmation of abnormal expression of the corresponding protein detected via IHC staining.
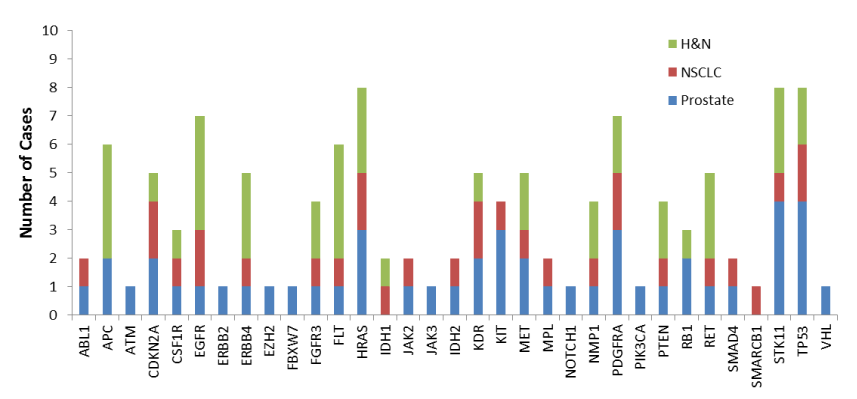
Figure 1.Gene specific occurrence of mutations in ROIs of specimens evaluated with the targeted Cancer Hotspot Panel from Thermo Fisher. Analysis included 4 prostate cancer specimens, 3 NSCLC specimens, and 4 H&N squamous cell carcinoma specimens
Three prostate cancer specimens that were heterozygous for loss of PTEN were annotated and isolation of tumor regions positive for PTEN were dissected and sequenced. One case was found to have 20 SNPs, including SNPs in ERBB2 (FOV = 4.3%) and EGFR (FOV = 36.4%), in the ROI. These mutations were not identified when the whole tissue specimen was sequenced. Subsequent staining of the prostate specimen for HER2 and EGFR confirmed overexpression in the tumor ROI that was isolated and sequenced (Figure 2A). A second PTEN heterozygous prostate specimen was found to have 19 SNPs. A mutation in EZH2 was present (FOV = 3.4%) only in the ROI that was positive for PTEN. Staining confirmed overexpression of EZH2 in the tumor ROI that was isolated and sequenced (Figure 2B). A third PTEN heterozygous prostate specimen had 12 SNPs, including multiple mutations in KIT (FOV = 3.2%, 15.2%) in the ROI. Staining confirmed abnormal expression of KIT in the tumor ROI (Figure 2C).
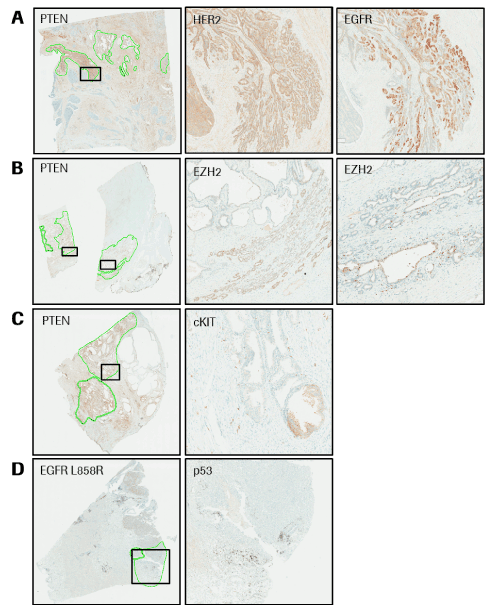
Figure 2.Case examples from Integrated CDx Workflow. Highlighted areas represent annotated tumor regions that were excised and sequenced. (A) Prostate cancer specimen stained with anti-PTEN primary antibody and displaying heterogeneous staining. Highlighted ROI is positive for abnormal HER2 and EGFR. (B) Prostate cancer specimen stained with anti-PTEN primary antibody and displaying heterogeneous staining. Highlighted ROI is positive for abnormal EZH2. (C) Prostate cancer specimen stained with anti-PTEN primary antibody and displaying heterogeneous staining. Highlighted ROI is positive for abnormal cKIT. (D) NSCLC specimen stained with anti-EGFR L858R mutation and displaying heterogeneous staining. Highlighted ROI is negative for presence of p53.
A lung cancer specimen that was heterozygous for the EGFR L858R mutation was annotated and the tumor regions negative for the EGFR mutation was dissected and sequenced. The case was found to have 26 SNPs, including 7 SNPs in TP53, 6 of which were not present when sequencing the whole specimen (FOV = 1.7% to 35.7%). Staining confirmed that the whole tumor had lost p53 protein expression (Figure 2D).
In this proof-of-principle study, actionable biomarkers were successfully identified in 70% of tumor cases utilizing our ICDx approach. Additionally, identification of HER2 and EGFR over-expression in a prostate cancer specimen (Figure 2A) is the strongest example of verifying actionable markers with clinically validated IHC tests with potential for directing treatment. These tests determined potential candidate biomarkers based on data derived from sequencing results. Using this integrated companion diagnostic approach, the patient could have been treated with anti-EGFR and/or anti-HER2 drugs, which would have been overlooked with conventional methodologies. Further studies, with increase sample numbers, would be necessary to confirm theories.
The importance of sequencing specific ROIs to identify potentially actionable mutations is demonstrated by the loss of specific mutations when a whole specimen is sequenced compared to tumor ROIs. Overall, we found that the use of tumor specific isolation in regions of interest allowed for detection of mutations that would otherwise go undetected in the majority of cases, several of which were clinically actionable. Isolation of these ROIs was made possible by use of automated dissection that allowed for precision detection and a standardized process.
With over 300 IHC assays available in our CAP/CLIA accredited laboratory, it is faster to screen a specimen with a targeted sequencing panel to identify potential markers of interest that may be contributing to heterogeneous cancer formation. While the majority of mutations identified via sequencing do not result in corresponding abnormal protein expression, we have shown that sequencing data can be successfully used to suggest potential biomarkers of interest. This is in addition to the general knowledge gained from sequencing to help characterize the tumor on a molecular basis, particularly in regards to tumor heterogeneity.
Authorship: HG performed extractions, sequencing, and data analysis. NT performed IHC staining and analysis. HG drafted the manuscript. All authors read and approved the final manuscript.
Funding: Research was supported by private funds.
Competing interest: All authors are employees for Ventana Medical Systems, Inc, a member of the Roche group.
2021 Copyright OAT. All rights reserv
- Welch DR (2016) Tumor Heterogeneity--A 'Contemporary Concept' Founded on Historical Insights and Predictions. Cancer Res 76: 4-6. [Crossref]
- Heppner GH (1984) Tumor heterogeneity. Cancer Res 44: 2259-2265. [Crossref]
- Marusyk A, Polyak K (2010) Tumor heterogeneity: causes and consequences. Biochim Biophys Acta 1805: 105-117. [Crossref]
- Hardiman KM, Ulintz PJ, Kuick RD, Hovelson DH, Gates CM, et al. (2016) Intra-tumor genetic heterogeneity in rectal cancer. Lab Invest 96: 4-15. [Crossref]
- Maley CC, Galipeau PC, Finley JC, Wongsurawat VJ, Li X, et al. (2006) Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet 38: 468-473.
- Taniguchi K, Okami J, Kodama K, Higashiyama M, Kato K (2008) Intratumor heterogeneity of epidermal growth factor receptor mutations in lung cancer and its correlation to the response of gefitinib. Cancer Sci 99: 929-935. [Crossref]
- Aran D, Sirota M, Butte A (2015) Systematic pan-cancer analysis of tumor purity. Nat Comm 6: 8971.
- Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, et al. (2007) Molecular definition of breast tumor heterogeneity. Cancer Cell 11: 259-273. [Crossref]
- Macintosh CA, Stower M, Reid N, Maitland NJ (1998) Precise microdissection of human prostate cancers reveals genotypic heterogeneity. Cancer Res 58: 23-28. [Crossref]


