Increased levels of cholesterol and saturated fatty acids (FAs) in immune cell membranes have proinflammatory effects. As opposed to specific effects of lipids mediated by certain lipid-protein interactions, non-specific and indirect effects of such lipids that regulate protein dynamics, regardless of their sequence, could have played influential roles in the early stages of evolution of life. Our recent atomistic simulations showed that, compared to bilayers with abundant unsaturated acyl chains, raft-like bilayers rich in cholesterol and saturated FA chains exert an effect to stabilize the dimeric state of transmembrane helical peptides with simple sequences. The energy cost associated with desolvation of the peptides from lipids upon dimerization was less in the raft-like bilayers compared to unsaturated FA-rich bilayers, suggesting that solvation (or fitting) of peptides by lipids is important for the dimer-stabilizing effects of such bilayers. In our simulations, acyl chains of phospholipids, but not cholesterol, directly solvated peptides. It is hypothesized that the peptide dimer-stabilizing effect may be the origin of the pro-inflammatory effects of cholesterol and saturated FAs. In this commentary, we mainly discuss our observations in atomistic simulations with some considerations on related experiments and computations as well as on recent knowledge on the properties of various membrane microdomains.
The effects of cholesterol and saturated fatty acids (FAs) have become widely recognized and imply the existence of complex interactions. Proinflammatory effects of these lipids species are considered to involve multilayered and multifaceted cooperative interactions of molecules involving proteins that can recognize specific lipid species [1-4]. On the other hand, as alterations of concentrations of these lipids can change the physicochemical properties of membranes, it is plausible that these lipids impact protein functions thorough mechanisms that are not dependent on specific amino acid sequences of proteins, i.e., in a sequence-nonspecific manner. So, when one questions biochemists or immunologists about possible mechanisms for the proinflammatory effects of lipids, responses are destined to be diverse, depending on the researchers' particular expertise.
Similar to, but slightly different from, the 'specific versus nonspecific' division is the classification of 'direct' and 'indirect' effects of particular lipids. 'Indirect' effects correspond to those situations in which particular types of lipids impact membrane protein structure/functions indirectly by modifying the bilayer environment, including its stiffness, fluidity, thickness, trans-bilayer hydrophobic mismatch, curvature, and lipid domains [5]. Such 'indirect' effects should basically be classified as 'non-specific' effects. While 'specific' effects should be largely mediated by 'direct' association of cholesterol or particular FAs with proteins, 'non-specific' effects may involve cases with diverse degrees of lipid-protein association, ranging from temporary contacts to relatively stable, prolonged binding. For example, 'tilted peptides' may be considered to have the ability to bind to cholesterol (enabling direct effects), defying sequence-based definition (nonspecific effects) [6]. We would like to refer readers to Lange and Steck [5] for diverse modalities of cholesterol-protein interactions.
For both cholesterol and FAs, recent efforts have uncovered important instances of specific and direct effects. As an example, for specific effects of FAs, n-3 PUFAs (DHA and EPA), but not saturated FAs, have been shown to exert potent anti-inflammatory effects through GPR120 [7]. In the case of cholesterol, well-studied peptide motifs that recognize it involve the CRAC motif ((L/V)-X1-5–(Y)-X1-5-(K/R)) and the CARC motif, which is similar to CRAC, but has an opposed orientation [6]. Other motifs involve the sterol-sensing domain (SSD), which has been identified in several enzyme and transport proteins important for cholesterol metabolism [5]. To our knowledge, the biological significance of these motifs is largely elusive. Nevertheless, more examples of specific cholesterol effects are likely to be elucidated in the future. In the case of the β-2 adrenergic receptor, cholesterol is likely to facilitate interaction between G protein-coupled receptors (GPCRs) [8]. Hanson and colleagues presented an X-ray crystallography model of human β-2 adrenergic receptor in which cholesterol is accommodated in the groove formed by four transmembrane (TM) α-helices [9] Using atomic force microscopy-based single-molecule force spectroscopy, Zocher et al. [10] for example, showed that cholesterol (cholesteryl hemisuccinate was used) considerably augmented the strength of interactions stabilizing structural segments of the β-2 adrenergic receptor. Besides GPCRs, the ability of cholesterol to modulate activities of several ion channels and transporters, including Na+, K+-ATPase, is well-established, as discussed in Lange et al. [5]. For Kir2 and heterotetrameric Kir3.1/Kir3.4 channels, specific mutations abrogated cholesterol’s effects, supporting the concept of direct binding by cholesterol to a specific site of such channels [11]. Although in some cases (dopamine transporter of Drosophila melanogaster, and Na+, K+-ATPase from pig kidney), the cholesterol binding site was identified by crystallographic analysis, the involvement of indirect effects cannot be excluded from the observed physiological effects of cholesterol.
With respect to nonspecific effects, Matsuzaki and associates demonstrated that addition of cholesterol stabilized the dimeric state of (AALALAA)3 peptide [12] using liposomes composed of palmitoyloleoylphosphatidylcholine (POPC). However, to our knowledge, fewer studies have focused on nonspecific effects. Nonetheless, nonspecific effects may have been important as the basis that determines or influences the direction of long-term evolution, which favored pro-inflammatory roles for these lipids. In other words, nonspecific physicochemical effects might have influenced the 'evolutionary fate' of cholesterol and saturated FAs as proinflammatory factors; in this view, newly evolved molecules (proteins) may have augmented or fine-tuned such effects of these lipids by elaborating specific protein-lipid interactions.
Interactions between TM domains of membrane proteins have drawn researchers' interest from biomedical and pharmaceutical perspectives. Association between TM helices is a widely-used strategy that directs the assembly of protein complexes and mediates signal transduction of hormone and cytokine receptors. At least for many single-pass TM proteins, disease-associated mutations in TM domains have been reported. It seems convenient to tentatively consider that there are two broad categories of interactions [13]. The first involves static interactions, in which the TM domains form relatively fixed contacts necessary for the assembly of a functional protein complex and for proper folding of proteins. The second category is that of dynamic conformational changes, in which these changes are propagated through the membrane via changes in the oligomerization state and/or orientation of TM helices [13]. Of note, Toll-like receptor (TLR) activation is accompanied by dimerization of TM domains [14]. As a well-studied example, TM domains of TLR2 and 6 are known to be important for activation of these proteins, interacting with each other and forming heterodimers. An inhibition analysis using synthetic peptide from TLR2 TM domain supported this conclusion [15].
There is currently no universal consensus model for signaling mechanisms mediated by diverse single-span membrane proteins. In general, the dimeric state involves the active configuration/conformation. The simplest model ('canonical model') for the signaling of cytokine receptors and receptor tyrosine kinases was an equilibrium between inactive monomers and ligand-bound dimers [16,17]. However, recent studies have extended this view; a substantial portion of receptors is forming inactive pre-dimers and, in addition to dimerization, subtle conformational changes are likely necessary for activation ('pre-formed dimer model') [18,19]. In the case of epidermal growth factor receptor (EGFR), a well-studied example of receptor tyrosine kinases, inactive symmetric intracellular kinase domains need to change to enzymatically active (asymmetric) dimers for EGFR signaling [20-22]. For human vascular endothelial growth factor receptor (VEGFR)-2, which represents another family of receptor tyrosine kinases, 30 to 60% of it was shown to exist in the dimeric form in the absence of ligand based on a Fluorescence resonance energy transfer (FRET) analysis, and ligand binding induces a conformational change in the TM domain dimer structure causing increased phosphorylation and further structural changes [19]. Human growth hormone receptor (hGHr), which, like cytokine receptors, utilizes JAK kinase for phosphorylation of the cytosolic domain, has also been shown to assume an inactive dimer conformation. A scissor-like motion of the TM domain of hGHr can switch the conformation of the intracellular domains, coupled with JAK2, between the active and inactive states, whereas for both states the TM domain is dimerized [23]. Thus, although future analyses may elucidate more cases of inactive pre-dimers and switching among dimeric conformations regulating activation, in general, the understanding that dimerization/oligomerization of TM domains is prerequisite for receptor activation is likely to remain valid. The dimerization energy discussed in our study should govern the monomer-dimer equilibrium, influencing the density of inactive pre-dimers and, indirectly, that of the active dimers.
Switching between dimeric and monomeric states, as well as switching among different conformations of dimer states, is considered to be enabled by rather weak interactions via van der Waals interactions, as opposed to involving strongly polar or charged residues. In fact, for a number of activating mutations, replacement with Cys, Asp, Glu and Arg residues enhances strong electrostatic attraction, which could lock the protein in active conformations [13].
There have been coarse-grained (CG)-based analyses on dimerization/clustering of TM peptides in phospholipid membranes [24-26]. However, CG models typically represent four hydrocarbon chains of (CH2)4 with one particle; in addition, Lennard-Jones (LJ) and Coulombic (electrostatic) energies have limited accuracies compared to atomistic models. In simulations, LJ energy represents the attractive force (due to van der Waals interactions) plus short-range repulsion. Therefore, it is generally difficult to gain insights from CG simulations into the mechanisms of atomic details for the effects of lipids on TM peptide behavior. We have been focusing on atomistic simulations to study mechanisms by which FAs and cholesterol control protein clustering. To our knowledge, no free energy analysis of cholesterol and/or FA effects on TM using popular all-atom parameters, such as CHARMM [27,28], AMBER [29] and Slipids [30], has been reported. From our experience involving cholesterol-containing bilayers, a simulation to achieve resolution at about ± 1 kJ/mol in energy analyses with all-atom parameters is likely to be a cost-intensive computation (e.g., 200 CPUs for one month). Hence, we took advantage of the GROMOS [31] united-atom model that represents CH2 and CH3 of carbohydrate chains as individual particles, achieving the precision of ~1 kJ/mol with thirty ordinary 4-core CPUs for two months. This allowed us to compare DOPC and raft-like bilayers in poly-Ile peptide self-dimerization [32].
Strikingly, cholesterol- and saturated FA-containing bilayers (a raft-like 1:1:1 POPC/DPPC/cholesterol bilayer, which we henceforth call 'the 1:1:1 bilayer') exhibited the dimerization energy of −2.23 kJ/mol for the self-associated state of the TM helical peptide poly-Ile ((I)21), which was 2.22 kJ/mol lower than the value (−0.01 kJ/mol) obtained for the DOPC bilayer. Thus, the presence of cholesterol and saturated FA chains shifted the monomer-dimer equilibrium toward the dimeric state. Similar stabilization by the 1:1:1 bilayer was seen for another model peptide I(VI)10, (i.e., a 21-residue peptide with alternating Ile and Val), strongly suggesting that the stabilizing effect is sequence-nonspecific [our unpublished data]. Although the quantitative relationship between cholesterol and saturated FA concentration and the degree of their stabilizing effects on dimerization is difficult to address, our preliminary analysis with the poly-Ile peptide with a 2:1:1 POPC/DPPC/cholesterol bilayer has shown a lesser degree of stabilization of the dimeric state compared to that of the 1:1:1 bilayer [our unpublished data]. This points to a perspective that the higher the order parameter of the lipid acyl chains is, the stronger the stabilizing effect on the dimer becomes.
The computational finding of saturated FA chain- and cholesterol-dependent shift of the monomer-dimer equilibrium toward the dimeric state corroborated the experimental findings by Yano et al. 2015 that showed that at 298 K dimerization free energy ∆Gmon->dim of (AALALAA)3 peptide was changed from −13.2 (POPC) to −22.6 kJ/mol (7:3 POPC:cholesterol), indicating stabilization of ~9 kJ/mol [12]. Of note, at 318 K, ∆Gmon->dim was −7.5 kJ/mol [12]. Our computation showed use of 1:1:1 bilayer led to −2.2 kJ/mol stabilization compared to DOPC bilayer, at 323 K, in a qualitative agreement with this experiment but exhibiting a significantly smaller difference. This could be due to peptide difference (Yano et al. [12] used (AALALAA)3, whereas we used a poly-Ile peptide) and definition of dimerized state (we regarded peptides > 1.6 nm distant from each other as the monomeric state), and possibly inaccuracy of the force field used. Of note, we compared DOPC and 1:1:1 bilayers, and Yano et al. compared POPC and 7:3 POPC/cholesterol bilayers. Although explicit replacement of unsaturated with saturated FAs was not tested in Yano et al. [12], the addition of cholesterol should dramatically increase the order parameter of acyl chains (i.e., straightening acyl chains). Our data also showed that cholesterol molecules contacted directly with phospholipids but not with peptides, with most cholesterol molecules residing > 0.3 nm distant from the peptides’ surfaces, whereas both POPC and DPPC directly contacted the peptides (Figure 3 of ref 32). Therefore, in the case of Yano et al. [12] as well, it is most likely that the effects of cholesterol are indirect; phospholipid molecules whose structures are modified by cholesterol are likely to directly stabilize the dimeric state of peptides.
As we have seen above, our computation supported the view that cholesterol and saturated FA acyl chains in bilayers stabilize the dimerized state of peptides. A key question is how they do so. Prior to considering potential mechanisms, we may be reminded of a few terms that describe energies. The free energy change ∆G is comprised of two components: the enthalpy change ∆H and the entropy change –T∆S. Hence, the free energy change upon transition from the monomeric to the dimeric state ∆Gmon->dim is decomposed into the two terms, i.e., ∆Gmon->dim = ∆Hmon->dim – T∆Smon->dim. In our case, the enthalpy ∆H is further decomposed into ∆U + P∆V, where U is the total internal energy (the sum of the kinetic and potential energies), and P∆V is the pressure-volume term, which is usually small. So, the enthalpy change ∆H becomes largely attributable to the change in the total internal energy term ∆U, which in turn parallels the total potential energy term ∆VLJ+Coul (that is, the sum of LJ and Coulombic potential energies) under constant temperature simulations. So, ∆Hmon->dim can be approximated by ∆VLJ+Coulmon->dim
Yano et al. 2015 showed that dimerization in POPC is an enthalpy-driven process (∆Gmon->dim = –13.2 kJ/mol, ∆Hmon->dim = –23.7 kJ/mol, –T∆Smon->dim = 10.4 kJ/mol) and that the addition of cholesterol to the POPC bilayer increased the unsigned value of ∆Gmon->dim and ∆Hmon->dim, thus stabilizing the peptide dimerization (∆Gmon->dim = –22.6 kJ/mol, ∆Hmon->dim = –84.1 kJ/mol, and –T∆Smon->dim = 61.4 kJ/mol) [12]. Thus, the addition of cholesterol increased the enthalpy differential ∆H between the monomers versus dimer states. The entropy change –T∆S was inhibitory to dimerization and was even more so in the presence of cholesterol [12]. In our simulations with the 1:1:1 bilayer, ∆Hmon->dim upon dimerization was approximately 3 kJ/mol for DOPC, and this value dropped to -25 kJ/mol in the raft-like bilayer, based on the comparison between r = 1.3 (dimer) and 1.6 nm (monomers). Thus, although the aforementioned differences between the experimental and computational approaches may have resulted in this wide discrepancy, both the approaches revealed similar trends; that cholesterol (cholesterol/saturated FA) induced stabilization of the dimeric state is an enthalpy-driven change.
Thus, although we ignore the entropy changes and the effect of the umbrella constraints on the potential energy that may have between–window variances, it is possible to discuss the dimerization free energy in terms of the potential energy terms between the peptides. This provides an advantage because the total potential energy ∆VLJ+Coul can be decomposed further. The total potential energy relevant to peptide dimerization is mainly comprised of three major terms-the (specific) peptide-peptide term, the lipid-peptide term, and the lipid-lipid term. (We avoid the word ‘specific’ henceforth to avoid confusion when we refer to them) [25,33]. It is often useful to consider the solvation of the peptides by lipids. In our case, ‘poor solvation’ means the peptide-lipid interaction that has the peptide-lipid potential energy term remaining at a large value (and likely causing high free energy) whereas a ‘good (or ‘better’) solvation’ indicates favorable peptide-lipid interactions resulting in a low peptide-lipid potential energy term (and likely resulting in low free energy). To be stable points to a low peptide-lipid potential energy term (i.e. ‘favorable’).
Let us consider plausible mechanisms by which cholesterol and saturated FA chains can stabilize the dimeric state of peptides. 1) In the first scenario, raft-like compositions may lead to tight lipid-lipid interactions, and such interactions might expel peptides from cholesterol and saturated FA (Figure 1A), stabilizing the dimeric state ('segregation-based mechanism'). 2) In the second scenario, solvation might remain ‘not so compromised’ when the peptides dimerize in the raft-like bilayer compared to the case in the DOPC bilayer ('solvation-based mechanism') (Figures 1B & 1C). In other words, while dimerization is always unfavorable to lipid-peptide interactions (as peptide dimerization always expels some lipids from the peptide-peptide interface) and increases the potential energy as the peptides approach each other (as represented by the ‘DOPC’ curve of Figure 1C), some properties of raft-like bilayer might somehow help solvate the dimerized peptides and diminish the lipid-peptide term of the potential energy (the 'raft-like' curve of Figure 1C). 3) In the third scenario, the protein-protein potential energy is the key factor. That is, tilt and cross angles differ depending on the lipid composition (raft-like understandably leads to upright orientations, with both angles small), and we need to examine if this feature influences peptide dimerization. However, 3) is unlikely, as the peptide-peptide interaction potential energy was comparable between DOPC and the raft-like bilayer (Figures 4A and 4D of ref 32).
We initially considered that 1), i.e., the segregation-based mechanism, might be the main factor (Figure 1A); this may increase the local concentration of peptides (i.e., peptide concentration within the cholesterol-low compartment), increasing the chance to form dimers. Under this scenario, we anticipated that the lipid-lipid potential energy term should support dimerization through decreasing (stabilizing) the term when the monomeric state changes to the dimeric one. However, our decomposition analysis argued against the concept that the effect of cholesterol that causes a more rigid property of raft bilayers plays a key role. Unexpectedly, the lipid-lipid interaction potential energy profile did not display a pattern in favor of peptide dimerization (Figures 4C and 4F of ref 32). Although the lipid-lipid potential term decreased upon dimerization as expected, the diminution for the raft-like membrane was somewhat less pronounced relative to that for the DOPC bilayer. Unexpectedly, scenario 2) was seen to be the case. This conclusion was supported by our additional analysis using the I(VI)10 peptide with the 1:1:1 bilayer as well as by that using the poly-Ile peptide with the 2:1:1 bilayer.
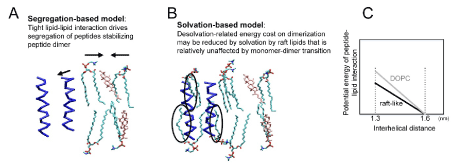
Figure 1. Plausible accounts for cholesterol and saturated FA-induced stabilization of the dimeric state of TM helical peptides. (A) Segregation-based model. In raft-like bilayers, close association between cholesterol and phospholipids may cause exclusion of TM peptides, increasing the local density of the peptides and the probabilistic likelihood of dimerization. (B) Solvation-based model. Peptide dimerization is invariably unfavorable to the lipid-protein interaction potential energy term because dimerization obstructs regions of peptide surfaces from solvation by lipids. However, in some mechanisms, dimerized peptides may be better solvated by lipids in raft-like bilayers, compared with instances involving unsaturated acyl chain-rich bilayers. (C) Schematic graphic representation of our recent results, which support the solvation-based model.
Our results add to a growing body of evidence that reveals that increases in cholesterol composition in lipid bilayer membranes stabilize the dimeric state of TM helical peptides [12]. However, phospholipids, but not cholesterol, directly contacted the peptides (< 0.3 nm) in our simulations. Further, our decomposition analysis showed that solvation of peptides by lipids is the key factor that causes enhanced stability of the dimeric state of peptides in the 1:1:1 bilayer. We surmise that the cholesterol elevates the order parameter of the phospholipid acyl chains that are directly in contact with the peptides, and that these straightened acyl chains lead to consequent improvement in the solvation of peptides, particularly when the dimer is formed.
Experiments using lipid vesicles that have mixtures of Ld and Lo domains showed that, in general, TM peptides are excluded from Lo domains [34,35], as their inclusion causes an energy cost required to break tight lipid associations in Lo domains. Similar exclusion was also seen by Baumbart et al., who used giant plasma membrane vesicles (GPMVs) [36], whereas results from Lingwood et al.’s plasma membrane spheres (PMSs) indicated a selective inclusion of raft TM proteins [37]. Thus, in general, Lo microdomains are not the favored locales for peptide partition compared to Ld microdomains. Schäfer et al. [33] showed that lipid packing drives the segregation of TM helices into disordered lipid domains in model membranes. In the latter study, both CG simulations and confocal microscopy-based experiments showed partition of WALP (Trp-flanked poly-(Leu-Ala) stretch of variable length) helical peptides into Ld phases. In the CG system, this energy is considerable-60 kJ/mol [33]. These findings corroborate with the discussion by Lorent and Leventhal [38] that palmitoylation is an important determinant for protein partition into lipid rafts, and that no, or few, other factors that unarguably act to drive lipids into rafts are recognized. Interestingly, our atomistic simulations of 1:1:1 composition revealed that cholesterol seldom contacted directly with peptides, likely because cholesterol-DPPC (POPC) is very favorable enthalpically, and disruption of this by peptides is not favored.
Then, one may ask, why have lipid rafts become utilized during evolution as a platform for protein clustering and signal transduction? We note that partition analyses compare partition but do not address the dimeric versus monomeric states. As our analyses showed, dimerization per se may be stabilized in lipid rafts. It should also be noted that in realistic systems, raft and non-raft exhibit very much smaller differences. Kaiser et al. [24] used two distinct types of plasma membrane-derived vesicles, GPMVs and PMSs. Although both GPMV and PMS exhibit separation of micro-meter sized phases, a lipid packing analysis using Laurdan and C-Laurdan by Kaiser et al. [24] showed that both GPMVs and PMSs show generally small differences between the ordered and less ordered phases. (They also demonstrated that the raft phase of PMS is less ordered than that of the Lo phase.) Such similarity between phases suggests little free energy differences in terms of partition, as well. Acylation of proteins should further lower the free energy of the partition into ordered microdomains. Future studies may involve drawing complete thermodynamic circles for monomeric versus dimeric states, as well as residing within versus outside of lipid rafts, focusing on more realistic systems. Atomistic simulations may be beneficial in this regard as solvation energy is difficult to discuss in CG simulations.
Hydrophobic mismatch effects could be a potential confounding factor. In theory, it is important to consider the mismatch concept because it is possible that raft-mediated cell signaling may utilize this aspect. However, we surmise that mismatch effects are not influential in our setting for the following reasons. First, our DOPC bilayer was as thick as our raft-like bilayer (with the distance between two layers of phosphorus atoms being 4.3 nm for both DOPC and the 2:1:1 bilayer), and it is unlikely to have exerted significant mismatch effects in our case, because the more upright orientation of the peptides in the raft-like bilayer would reduce the likelihood of negative mismatches. The positive mismatch penalty is generally minor due to facile accommodation of tilted peptides. Second, unlike our recent studies in which peptides were capped with polar or charged residues (Ser or Lys), we did not employ such caps in our 1:1:1 bilayer analysis.
Is peptide tilt angle a key factor contributing to dimer stability? Our comparison between the DOPC and the raft-like membranes showed that the latter bilayers favored lesser-tilted angles of the helices compared to the DOPC bilayers, but the difference was small. In fact, in the DOPC bilayers, the helices assumed tilt angles of 11-24 degrees (average = 15.3º) from the z-axis (bilayer normal) whereas in the raft-like 2:1:1 bilayers these remained within 10-22º (average = 14.2º). In addition to this unexpectedly small difference, the finding that the protein-protein potential energy profile did not show features suggestive of this term and help stabilize peptide dimerization, argues against the notion that tilt angle itself is an important factor for the stabilization of the dimeric state.
To summarize, our atomistic simulations revealed that cholesterol and saturated FA chains stabilize the dimeric state of helical TM peptides in an indirect and sequence-nonspecific manner. Decomposition analysis demonstrated that lipid-peptide interaction potential energy is the key factor contributing to stabilization of the dimeric state. As cholesterol generally acts to straighten the FA chains of phospholipids, and as phospholipids, but not cholesterol, directly contacted the peptide dimer, we surmise that structures of phospholipids in raft-like membranes offer some functional advantage in stabilizing peptide dimers. Consistent with this view, our preliminary comparison between DOPC and POPC bilayers indicated increased stability of peptide dimers in the POPC compared to the DOPC bilayers (our unpublished result). Hence, even without cholesterol, saturated FAs alone are likely to exert a stabilizing effect on peptide dimers. Whereas some authors have discussed that presence of cholesterol causes 'lipophobic effects' that lead to stabilization of the dimeric state, our simulation data pointed to a view that, rather, cholesterol enables phospholipid-protein interaction (that is, protein solvation by phospholipids). We propose that solvation, which is maintained relatively well, even after peptide dimerization, is likely to stabilize dimers compared to the case without cholesterol.
However, there remain several issues to be addressed in future studies. First, there remains a broad gap in our argument-our hypothesis is based on the premise that cholesterol- and saturated FA-induced stabilization of dimers/oligomers of TM peptides should augment the general tone of inflammation toward higher degrees of inflammation. This issue may be difficult to address experimentally, and therefore comprehensive considerations regarding molecular evolution of inflammation in various organisms would be necessary. Second, apart from practical biological issues, our analysis may have suffered from technical challenges; we need to expand the range of sampling because we mainly used inter-helical separations of 1.3 and 1.6 nm to evaluate the dimer and monomer states, respectively, whereas involvement at 2.5 nm is likely more desirable for more rigorous assessment [39,40]. Third, while united-atom parameters allow rapid convergence, our results need to be verified using all-atom force fields. Fourth, it seems important to ask whether the effects of cholesterol can be observed for a wider range of order parameters. More specifically, we speculate whether the peptide dimer stabilization we observed is a phenomenon which depends on the presence of a particular microdomain as a prerequisite, or whether, for a broad range of bilayers covering Lo and Ld bilayers, cholesterol exhibits similar effects. This is important because the existence of lipid rafts is controversial in many settings [5], and, in this sense, robustness of the cholesterol and saturated FA effects would liberate the discussion from controversies involving rafts. Our preliminary observation of the POPC > DOPC difference in the dimer stabilization effect is supportive of the dimer stabilizing effects for a wide range of bilayers, but further comparison would provide additional insights. Fifth, for physiological discussions, it is also important to examine the effect of asymmetry in lipid composition between two monolayers. Finally, transferability of our results involving model peptides with simple sequences to more biologically relevant systems should be addressed. In a more biological context, the relative impact of specific interactions between peptides and particular lipids might be more influential, so quantitative analysis becomes increasingly important. Furthermore, in realistic systems, mismatch- and tilt-angle effects may exert different degrees of impact, depending on the specific structures of the relevant peptides.
From a technical perspective, it is encouraging that at least united atom-level analyses of dimerization energies to a precision of 1 kJ/mol is feasible using 30 CPUs in a matter of months. Such resolution would enable analyses of TM domains of receptors that address, for example, effects of certain amino acid sequences that may be important to fine-tune the strength of interactions. It currently appears problematic to explain the atomistic reasons for how acyl chains with high-order parameters become favorable for solvation of peptide dimers compared to those with low-order parameters. Further detailed analyses focusing on the impact of tilt angles of TM helical peptides, and on solvation of dimers of a variety of peptides, may provide further mechanistic and structural clarity.
- Calder PC (2008) The relationship between the fatty acid composition of immune cells and their function. Prostaglandins Leukot Essent Fatty Acids 79: 101-108. [Crossref]
- Estadella D, da Penha Oller do Nascimento CM, Oyama LM, Ribeiro EB, Dâmaso AR, et al. (2013) Lipotoxicity: effects of dietary saturated and transfatty acids. Mediators Inflamm 2013: 137579. [Crossref]
- Kleemann R, Verschuren L, van Erk MJ, Nikolsky Y, Cnubben NH, et al. (2007) Atherosclerosis and liver inflammation induced by increased dietary cholesterol intake: a combined transcriptomics and metabolomics analysis. Genome Biol 8: R200. [Crossref]
- Ruiz-Núñez B, Dijck-Brouwer DA2, Muskiet FA2 (2016) The relation of saturated fatty acids with low-grade inflammation and cardiovascular disease. J Nutr Biochem 36: 1-20. [Crossref]
- Lange Y, Steck TL (2016) Active membrane cholesterol as a physiological effector. Chem Phys Lipids 199: 74-93. [Crossref]
- Fantini J, Barrantes FJ (2013) How cholesterol interacts with membrane proteins: an exploration of cholesterol-binding sites including CRAC, CARC, and tilted domains. Front Physiol 4: 31. [Crossref]
- Oh DY, Talukdar S, Bae EJ, Imamura T, Morinaga H, et al. (2010) GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell 142: 687-698. [Crossref]
- Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, et al. (2007) High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science 318: 1258-1265. [Crossref]
- Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, et al. (2008) A specific cholesterol binding site is established by the 2.8 A structure of the human beta2-adrenergic receptor. Structure 16: 897-905. [Crossref]
- Zocher M, Zhang C, Rasmussen SG, Kobilka BK, Müller DJ (2012) Cholesterol increases kinetic, energetic, and mechanical stability of the human ß2-adrenergic receptor. Proc Natl Acad Sci USA 109: E3463-3472. [Crossref]
- Bukiya AN, Osborn CV, Kuntamallappanavar G, Toth PT, Baki L, et al. (2015) Cholesterol increases the open probability of cardiac KACh currents. Biochim Biophys Acta 1848: 2406-2413. [Crossref]
- Yano Y, Kondo K, Kitani R, Yamamoto A, Matsuzaki K (2015) Cholesterol-induced lipophobic interaction between transmembrane helices using ensemble and single-molecule fluorescence resonance energy transfer. Biochemistry 54: 1371-1379. [Crossref]
- Moore DT, Berger BW, DeGrado WF (2008) Protein-protein interactions in the membrane: Sequence, structural, and biological motifs. Structure 16: 991-1001. [Crossref]
- Reuven EM, Fink A, Shai Y (2014) Regulation of innate immune responses by transmembrane interactions: lessons from the TLR family. Biochim Biophys Acta 1838: 1586-1593. [Crossref]
- Fink A, Reuven EM, Arnusch CJ, Shmuel-Galia L, Antonovsky N, et al. (2013) Assembly of the TLR2/6 transmembrane domains is essential for activation and is a target for prevention of sepsis. J Immunol 190: 6410-6422. [Crossref]
- Schlessinger J (1988) Signal transduction by allosteric receptor oligomerization. Trends Biochem Sci 13: 443-447. [Crossref]
- Williams LT (1989) Signal transduction by the platelet-derived growth factor receptor. Science 243: 1564-1570. [Crossref]
- Bocharov EV, Sharonov GV, Bocharova OV, Pavlov KV (2017) Conformational transitions and interactions underlying the function of membrane embedded receptor protein kinases. Biochim Biophys Acta: S0005-2736. [Crossref]
2021 Copyright OAT. All rights reserv
- Sarabipour S, Ballmer-Hofer K, Hristova K (2016) VEGFR-2 conformational switch in response to ligand binding. Elife 5: e13876. [Crossref]
- Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J (2006) An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 125: 1137-1149. [Crossref]
- Arkhipov A, Shan Y, Das R, Endres NF, Eastwood MP, et al. (2013) Architecture and membrane interactions of the EGF receptor. Cell 152: 557-569. [Crossref]
- Endres NF, Das R, Smith AW, Arkhipov A, Kovacs E, et al. (2013) Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell 152: 543-556. [Crossref]
- Brooks AJ, Dai W, O'Mara ML, Abankwa D, Chhabra Y, et al. (2014) Mechanism of activation of protein kinase JAK2 by the growth hormone receptor. Science 344: 1249783. [Crossref]
- Kaiser HJ, Orlowski A, Róg T, Nyholm TK, Chai W, et al. (2011) Lateral sorting in model membranes by cholesterol-mediated hydrophobic matching. Proc Natl Acad Sci U S A 108: 16628-16633. [Crossref]
- Castillo N, Monticelli L, Barnoud J, Tieleman DP (2013) Free energy of WALP23 dimer association in DMPC, DPPC, and DOPC bilayers. Chem Phys Lipids 169: 95-105. [Crossref]
- Prakash A, Janosi L, Doxastakis M (2011) GxxxG motifs, phenylalanine, and cholesterol guide the self-association of transmembrane domains of ErbB2 receptors. Biophys J 101: 1949-1958. [Crossref]
- Klauda JB, Venable RM, Freites JA, O'Connor JW, Tobias DJ, et al. (2010) Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B 114: 7830-7843. [Crossref]
- Lim JB, Rogaski B, Klauda JB (2012) Update of the cholesterol force field parameters in CHARMM. J Phys Chem B 116: 203-210. [Crossref]
- Madej BD, Gould IR, Walker RC (2015) A parameterization of cholesterol for mixed lipid bilayer simulation within the Amber Lipid14 force field. J Phys Chem B 119: 12424-12435. [Crossref]
- Jämbeck JP, Lyubartsev AP1 (2013) Another Piece of the Membrane Puzzle: Extending Slipids Further. J Chem Theory Comput 9: 774-784. [Crossref]
- Oostenbrink C, Villa A, Mark AE, van Gunsteren WF (2004) A biomolecular force field based on the free enthalpy of hydration and solvation: The GROMOS force-field parameter sets 53A5 and 53A6. J Comput Chem 25: 1656-1676. [Crossref]
- Nishizawa M, Nishizawa K (2017) Cholesterol and saturated fatty acid stabilize dimerization of helical transmembrane peptides by lowering energy cost related to peptides desolvation from lipids upon dimerization: an insight from atomistic simulation. Biomed Res Clin Prac 2: 1-8.
- Schäfer LV, de Jong DH, Holt A, Rzepiela AJ, de Vries AH, et al. (2011) Lipid packing drives the segregation of transmembrane helices into disordered lipid domains in model membranes. Proc Natl Acad Sci U S A 108: 1343-1348. [Crossref]
- Fastenberg ME, Shogomori H, Xu X, Brown DA, London E (2003) Exclusion of a transmembrane-type peptide from ordered-lipid domains (rafts) detected by fluorescence quenching: extension of quenching analysis to account for the effects of domain size and domain boundaries. Biochemistry 42: 12376-12390. [Crossref]
- Bacia K, Schuette CG, Kahya N, Jahn R, Schwille P (2004) SNAREs prefer liquid-disordered over "raft" (liquid-ordered) domains when reconstituted into giant unilamellar vesicles. J Biol Chem 279: 37951-37955. [Crossref]
- Baumgart T, Hammond AT, Sengupta P, Hess ST, Holowka DA, et al. (2007) Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc Natl Acad Sci U S A 104: 3165-3170. [Crossref]
- Lingwood D, Ries J, Schwille P, Simons K (2008) Plasma membranes are poised for activation of raft phase coalescence at physiological temperature. Proc Natl Acad Sci U S A 105: 10005-10010. [Crossref]
- Lorent JH, Levental I (2015) Structural determinants of protein partitioning into ordered membrane domains and lipid rafts. Chem Phys Lipids 192: 23-32. [Crossref]
- Nishizawa M, Nishizawa K (2014) Potential of mean force analysis of the self-association of leucine-rich transmembrane a-helices: difference between atomistic and coarse-grained simulations. J Chem Phys 141: 075101. [Crossref]
- Nishizawa M, Nishizawa K (2016) Free energy of helical transmembrane peptide dimerization in OPLS-AA/Berger force field simulations: inaccuracy and implications for partner-specific Lennard-Jones parameters between peptides and lipids. Mol Simul 42: 916-926.

