Introduction: Coronavirus crisis has activated healthcare administrations and personnel at highest level. Health informatics software tools and innovative e-governance policies can contribute to this point. In a previous study, we proposed a nationwide biomedical registry framework, that it could be valuable as a healthcare management tool of eGov Health, additionally.
Methods: Translating COVID-19 outbreak in health informatics terms, we developed virtual COVID- 19 datasets to demonstrate a generic COVID-19 patient recruitment system in Greece. An open- source JS library was also used to develop a custom interactive map in order to display valuable information for clinical trial researchers and crisis managers.
Results: Results indicate our proposed application enhances the clinical trial processes to recruit eligible patients as a clinical trial cohort. Registered physicians - clinical researchers can overview available COVID-19 patients per health unit, nationwide. The implemented virtual web maps provide a helpful visualization of COVID-19 crisis in Greece.
Conclusion: Our proposed recruitment system could function both as a PRS and a DSS tool under infection diseases parameters. We strongly believe our application has generic eGov potential, both as an enhancing tool to design a clinical trial and as a healthcare crisis management tool.
patient recruitment system, open-source software, COVID-19, clinical research, crisis management
COVID-19 outbreak is literally disrupting all human activities and is changing the modern stand of life in front of everyone’s eyes, reminding us about the tremendous consequences of previous pandemics at the same time [1,2]. Perspectives and potential pharmaceutical interventions continue filling the pages of many scientific journals. Generic prevention measures should always be considerable [3], but an effective vaccine or a therapeutic protocol for the disease is the immediate goal of biomedical laboratories at that time [4-7]. According to several global news agencies and publications, governments of advanced counties are pushing in that direction, thinking a possible virus mutation that it could lead to a COVID-20 outbreak, and so on [8-10].
Although healthcare employees and biomedical researchers are in the front of the struggle, coronavirus crisis is generating an enormous effort of information systems development and Internet exploitation, worldwide. Governments are rapidly implementing modern IT technologies for daily human social – economic activities, but caution must be taken concerning surveillance applications and person’s privacy in terms of effective containment measures [11-15]. As an extension of this fact, health IT managers, software developers and digital health start-ups have been already called to support new digital strategies and develop new digital solutions [16-18].
Parallel to most of the world, the healthcare crisis has serious implications in all human activities in Greece [19]. Public and private sector, schools and universities, banks, marketplaces, and enterprises are being forced to comply with web-based and cloud solutions [20-22]. From an IT standpoint, statistical e-Governance (eGov) indicators for Greece compared to the European Union average present an encouraging growth rate, according to the official Digital Governance Factsheets of 2019 for EU, but there is still a lot of room for improvement [13,23]. Thus, among the governmental social measures that have been taken to fight the COVID-19 crisis, a Hellenic National COVID-19 Patient Registry could be decided by the Hellenic Ministry of Health to be established. The Hellenic COVID-19 Register project could protect the population health in view of the great impact of the disease, the need to record epidemiological data, to record pharmacovigilance and the need for supervision of healthcare providers in the private sector. The registry might directly record the COVID-19 cases in all health units nationwide and to form sufficient epidemiological data for coronavirus, useful for following similar situations. Based on what is governmental approved, the registry database could record each COVID- 19 patient's name, age, gender, underlying diseases, and his/her state of health, under the mandatory GDPR compliance and web security techniques.
In a previous study in 2019, we introduced a virtual web-based national biomedical registry, named “HBR” (Hellenic Biomedical Registry), in order to enhance design conditions for clinical research in Greece, by integrating the clinical and molecular/genomic patient’s data [24]. Our proposed framework was based on WordPress® open-source software (FOSS) technology following the modern trends for agile and minimum cost of development. Moreover, we used colorectal cancer as an example to prove the functionality and the effectiveness of our software. In brief, the HBR web platform aims to help clinical medical doctors – researchers (1) to overview the stored integrated clinical and molecular/genomic data of submitted patients - volunteers, and (2) to query the total number of patients per disease/condition and simultaneously per health unit, nationwide. The system designed to function both as a dynamic Patient Recruitment System (PRS) and as an Electronic Biomedical Patient Record (EBPR).
From a health IT perspective, COVID-19 clinical research trigger our virtual system to demonstrate anew its functionality as an underlined PRS for a clinical trial process related to COVID-19. Applying the coronavirus disease ICD-10 as new parameter in HBR system instead of colorectal cancer, the key objective of the present manuscript is to call out the healthcare crisis managers that our software philosophy could cover the objectives of a National COVID-19 Patient Registry. Eventually, it could be an effective health Decision Support Software (DSS) tool, among others, in the fight against the crisis.
Managing our HBR application under the same configuration (as it was referenced in our previous published article), we briefly followed the next steps to re-evaluate the system perspectives in regard to coronavirus crisis: (a) we composed a virtual COVID-19 positive dataset of Greek patients, based on official ongoing COVID-19 information referred by the Hellenic National Public Health Organization [25], and we inserted it into our system database; (b) we preferentially used an open-source web mapping technology to geolocate our virtual COVID-19 data. Literature review was conducted in order to be up to date about the COVID-19 crisis management. Because of the profound COVID-19 crisis, it is mentioned that the World Health Organization provided information about additional ICD-10 codes for virus identified cases (U07.1) and virus non identified cases (U07.2) respectively [26]. These imbedded ICD-10 codes comprised fundamental parameters for our SQL queries.
In more practical detail, a data table was added into the HBR database with the geolocation of the leading Greek cities with the most of COVID-19 positive cases, according to governmental announcements until Apr 10th, 2020. Furthermore, another data table was intentionally added referring the aggregate number of virtual COVID-19 patients – willing volunteers per corresponding virtual health unit (Health Unit-A at Athens, Health Unit-B at Thessaloniki, Health Unit-C at Larissa), as potential participants in coronavirus-related clinical research. To embed source code for interactive map development, we handled Leaflet v.1.6.0 (https://leafletjs.com/, last accessed in April 10, 2020) as an open-source JavaScript library for web mapping, maintaining our principles for minimum cost of development. Connection strings with the HBR data sources were based upon the tutorial of the JavaScript library. Additional software implementation time was barely a week. Web maps source code is available upon request to our Laboratory including GeoJSON format. Figure 1 visualizes our methodology.
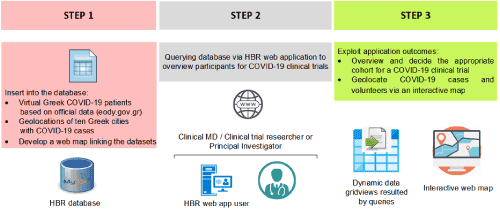
Figure 1. Approach to properly implement HBR as a DSS valuable tool in coronavirus crisis. Step 1: implement appropriate COVID-19 patient datasets into the HBR database and progressively develop a web map to display the inserted data. Step 2: registered users query the database through the web- based UI for specific COVID-19 information. Step 3: requested information is displayed through data grids and interactive maps
In the HBR system, ICD-10 is one of the required data elements which a registered MD should properly fill in the online electronic Case Report Form (eCRF) of his/her volunteer patient, as it is mentioned in our previous relevant publication for the Hellenic Biomedical Registry. In similar to previous colorectal cancer example, querying the HBR for standard COVID-19 ICD-10 codes in a virtual Health Unit ‘A’, the MD user gets the anticipated grid view of Figure 2(A). The data grid view provides dynamic sorting as well as editing grants according to HBR user essential role. Figure 2(B) displays the source SQL code for the accurate calculation of the number of COVID-19 cases per virtual health unit, by running multiple ‘select count’ SQL commands at the system’s backend, which eventually resulted to Figure 3(C). The displaying information of prospective COVID-19 patients provides a principal investigator/researcher the ability to rapidly create a prospective cohort of patients for an associated clinical trial.

Figure 2. Screenshots of the virtual HBR outcomes using COVID-19 as an example, based on realistic published data by Greek authorities. HBR ID format is used as primary key in data imports. Image (A) displays COVID-19 cases of three virtual health units. The data grid view provides dynamic sorting as well as editing grants according to HBR user’s role. Image (B) displays the source SQL code for the calculation of the number of COVID-19 cases. Image (C) shows the view of the previous multiple “Select Count” SQL command. Following the present methodology, each registered clinical researcher can overview the total number of coronavirus patients per health unit, nationwide, and as a result to be able to monitor the disease progression or design a cohort of cases around SARS-CoV clinical studies
Additionally, through the HBR data analytics web submenu items, we embedded a bar chart to display the disease progression of combined cases per leading city (Figure 3). The visualization of the direct comparison among distinctive regions could definitely lead official healthcare administrators to increase or to decrease control mechanisms of disease spread. Figure 4 displays a screenshot of the first developed interactive map, which virtualizes the aggregate number of COVID-19 cases by the comparable density of a color. A popup info box appears on region mouseover displaying relevant information. Similarly, Figure 5 shows a screenshot of the second implemented web map which is conformed to the new coronavirus ICD-10 codes. Therefore, each city - marker of the web map interacts on mouseover displaying a mini set of sufficient information about hospitalized COVID-19 cases, who could potential be considered as available patients for a clinical trial. As a result, registered MDs and clinical research researchers can accelerate their decisions regarding the establishment of an appropriate cohort of COVID-19 patients in the initial stages of clinical research design.
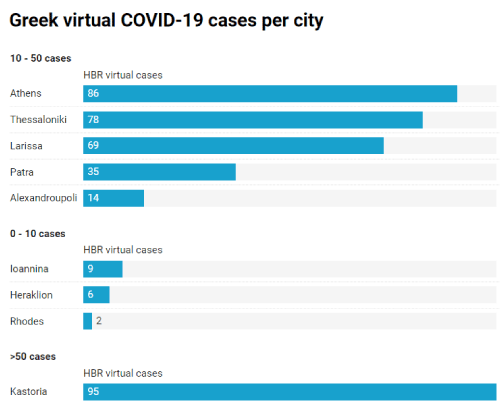
Figure 3. Screenshot of open-source developed bar chart in the HBR application, displaying the virtual COVID-19 patients per city (Based on factual report by official the Greek healthcare authorities April 10th, 2020, Eody.Gov.gr)
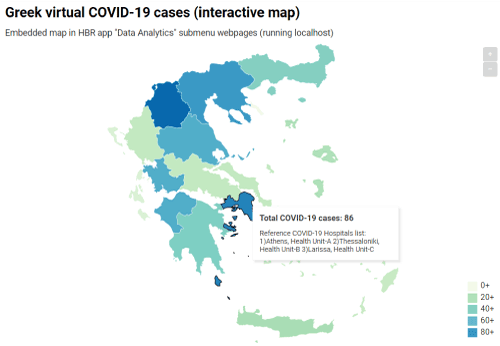
Figure 4. Screenshot of the embedded interactive Choropleth Map of Greece in the HBR application, displaying the virtual COVID-19 cases (Based on factual report by official the Greek healthcare authorities April 10th, 2020, Eody.Gov.gr). The comparable density of color regions depends on the number of cases. Each region mouseover emerges a popup info box
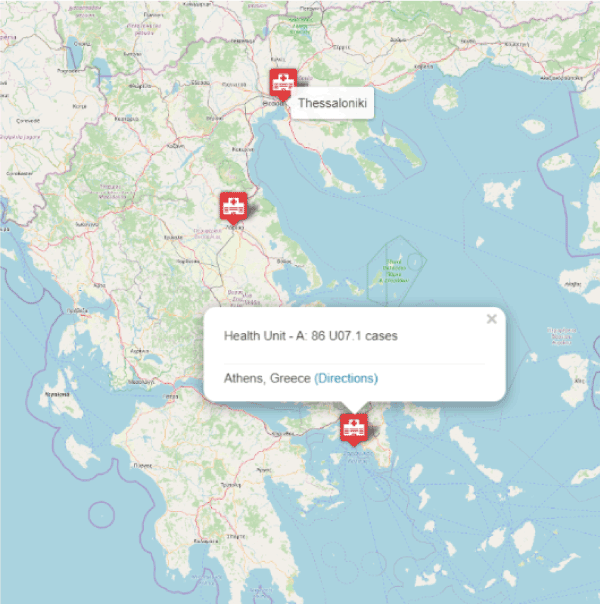
Figure 5. Screenshot of the embedded interactive Symbol Map of Greece in the HBR application, displaying the virtual COVID-19 cases (Based on factual report by official the Greek healthcare authorities April 10th, 2020, Eody.Gov.gr). A popup info box on each city - marker mouseover displays the total number of the cases per health unit and COVID-19 ICD-10 (U07.1 or U07.2)
Also, the promotion of biomedical collaboration among clinical researchers could be surprisingly enhanced while the HBR registered biomedical researchers could overview comprehensive information for a specific disease among hospitals nationwide.
It is more than certain, that the following years the eGov Factsheets and indicators will be more than satisfactory for all EU countries, because of the healthcare crisis. Hence, our proposed system could be implemented as a new eGov Health policy contributing to the improvement of e-governance healthcare in Greece.
Furthermore, by watching the World Health Organization to promote “Solidarity clinical trial for COVID-19 treatments” via its webpage [27], we are persuaded that every scientific or technological idea related to the treatment of a pandemic cause should be considerable by the healthcare crisis managers. The pressure COVID-19 puts on health systems should promptly lead Health Administrations to speed the adoption of health informatics proposals and the deployment of innovative software solutions. An addition of an official healthcare manager role with specific grants on the HBR application menu and views could be implemented through the system administration dashboard. Subsequently, the limited HBR registered healthcare administrators could exploit our system to take decisions on healthcare crisis progression.
Because of coronavirus outbreak, the efficient data sharing is undoubtedly becoming an emerging issue. Discussions for intense collaborations between Biomedical and health IT scientists in order to implement effective information system solutions and eGov policies may soon arise, due to the need of rapidly design and execution of infectious SAR-CoV viruses - related clinical research or to the need of a future similar crisis management [28]. At last, previous research has indicated that individual immune response variabilities in infection diseases (including SAR-CoV viruses) is regulated by host genomic factors variations, especially HLA genes polymorphisms [29-35]. Therefore, it could be advantageous for clinical physicians - researchers to conduct further clinical studies based on the exploitation of the easily further configured open-source HBR abilities to store molecular and genomic patient’s information.
From a web application security perspective, it must be emphasized that the personal data of the COVID-19 patients should be protected applying all necessary logical and physical anonymization techniques. But, information security measures are outside of the scope of our PRS demonstration at this time.
As overall conclusions, we believe that our previous published virtual biomedical registry covers the initial efforts to design and to conduct a clinical study under COVID-19 parameters. We have mentioned that the HBR application could literally work as a patient recruitment system as well as an intelligent decision support software tool for evaluating the potential of hospitalized patients or outpatients to participate in a research. In conclusion, we sincerely believe that our proposed PRS covers the requirements of a Hellenic COVID-19 registry, thus it could be easily implemented in the Greek eGov Health architecture and potentially in any eGov architecture, enhancing the recruiting processes of COVID-19 related clinical research.
No competing financial interests exists.
- Monto AS, Fukuda K (2020) Lessons from Influenza Pandemics of the Last 100 Years. Clin Infect Dis 70: 951–957.
- Rothan HA, Byrareddy SN (2020) The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun 109: 102433.
- Watkins J (2020) Preventing a covid-19 pandemic. BMJ 368: m810.
- Kalil AC (2020) Treating COVID-19-Off-Label Drug Use, Compassionate Use, and Randomized Clinical Research During Pandemics. JAMA 323: 1897-1898
- Kupferschmidt K, Cohen J (2020) Race to find COVID-19 treatments accelerates.
- Zhang T, He Y, Xu W, Ma A, Yang Y, et al. (2020) Clinical research for the treatment of Coronavirus disease 2019 (COVID-19): A rapid response to urgent need. Sci China Life Sci 29:1-3
- Cohen J (2020) Vaccine designers take first shots at COVID-19. Science 368: 14-16.
- Morissin JS, Carol A (2020) Which Covid-19 Future Will We Choose? Center for Strategic & International Studies.
- Knapton S (2020) Coronavirus has mutated into more aggressive disease, say scientists. The Telegraph.
- Pan X, Ojcius D, Gao T, Li Z, Pan C, et al. (2020) Lessons learned from the 2019-nCoV epidemic on prevention of future infectious diseases. Microbes Infect 22: 86-91.
- Bozorgmehr K, Saint V, Kaasch A, Stuckler D, Kentikelenis A (2020) COVID and the convergence of three crises in Europe. The Lancet Public Health 5: e247-248.
- Ting DS, Carin L, Dzau V, Wong TY (2020) Digital technology and COVID-19. Nature medicine 26: 459-461.
- Psaropoulos J (2020) Coronavirus accelerates Greece's overdue digital revolution. Al Jazeera [Internet].
- Valentino-DeVries J (2020) Translating a Surveillance Tool into a Virus Tracker for Democracies. The New York Times [Internet].
- European Commission Press corner (2020) State aid: Commission approves €2 billion Greek guarantee measure to support companies affected by the coronavirus outbreak [Internet].
- Reeves J, Hollandsworth H, Torriani F, Taplitz R, Abeles S, et al. (2020) Rapid Response to COVID- 19: Health Informatics Support for Outbreak Management in an Academic Health System. J Am Med Inform Assoc.
- Wind TR, Rijkeboer M, Andersson G, Riper H (2020) The COVID-19 pandemic: The ‘black swan’ for mental health care and a turning point for e-health. Internet Interv 20: 100317.
- Eccleston C, Blyth FM, Dear BF, Fisher EA, Keefe FJ, et al. (2020) Managing patients with chronic pain during the Covid-19 outbreak: considerations for the rapid introduction of remotely supported (e-health) pain management services. Pain 161: 889.
- Hellenic Government COVID-19 portal [Internet] 2020.
- National Documentation Center: EKT.gr. EITHealth Calling digital health start-ups with valuable COVID-19 solutions [Internet] 2020.
- Hellenic Ministry of Education: Eclass.Sch.gr (η-Τάξη) [Internet] 2020.
- Hellenic General Secretariat for Civil Protection: Forma.Gov.gr [Internet] 2020.
- NIFO - National Interoperability Framework Observatory. Digital Government Factsheets - 2019 [Internet] 2020.
- Kotoulas A, Lambrou G, Koutsouris DD (2019) Design and virtual implementation of a biomedical registry framework for the enhancement of clinical research: colorectal cancer example. BMJ Health Care Inform 26: 1-10.
- Hellenic National Public Health Organization: Eody.Gov.gr. Daily Reports COVID-19 [Internet] 2020.
- World Health Organization: Emergency use ICD codes for COVID-19 disease outbreak [Internet] 2020.
- World Health Organization: “Solidarity” clinical trial for COVID-19 treatments [Internet] 2020.
- Moorthy V, Restrepo AM, Preziosi MP, Swaminathan S (2020) Data sharing for novel coronavirus (COVID-19). Bulletin of the World Health Organization 98: 150.
- Chen YM, Liang SY, Shih YP, Chen CY, Lee YM, et al. (2006) Epidemiological and genetic correlates of severe acute respiratory syndrome coronavirus infection in the hospital with the highest nosocomial infection rate in Taiwan in 2003. J Clin Microbiol 44: 359-365.
- Blackwell JM, Jamieson SE, Burgner D (2009) HLA and infectious diseases. Clin Microbiol Rev 22: 370-385.
- Fitzmaurice K, Hurst J, Dring M, Rauch A, McLaren PJ, et al. (2015) Additive effects of HLA alleles and innate immune genes determine viral outcome in HCV infection Gut 64: 813–819.
- Tian C, Hromatka BS, Kiefer AK, Eriksson N, Noble SM, et al. (2017) Genome-wide association and HLA region fine-mapping studies identify susceptibility loci for multiple common infections. Nat Commun 8: 1-3.
- Sambaturu N, Mukherjee S, López-García M, Molina-París C, Menon GI, et al. (2018) Role of genetic heterogeneity in determining the epidemiological severity of H1N1 influenza. PLoS Comput Biol 14: e1006069.
- Kelly A, Trowsdale J (2019) Genetics of antigen processing and presentation. Immunogenetics 71: 161– 170.
- Sun Y, Xi Y (2020) Association Between HLA Gene Polymorphism and the Genetic Susceptibility of SARS Infection. Published 2014 March 19 [Internet].





