Angiogenesis, the growth of new blood capillaries from preexisting vessels, is a crucial process in the development and spread of cancers. The inhibition of angiogenesis thus appears as a promising therapy for cancer. Methionine aminopeptidase type 2 (MetAP2) is a metallo-enzyme considered today as an attractive target for the fight against cancer because its inhibition blocks the process of angiogenesis and therefore tumor progression. The aim of this work is to study the inhibition of MetAP II by molecular docking methods in order to discover new anti-angiogenic agents. FlexX is the molecular docking program used. Thus, this software allows predicting protein-inhibitor interactions. It was used to study the inhibition of 5JFR, a human MetAP II, by various molecules in order to discover the best inhibitors of this enzyme. The evaluation of the interaction energy of these inhibitors made it possible to identify those exhibiting the best inhibitory effect. These are compounds 6KP and A84, whose docking scores are -32.33 and -32.92 Kj/mol, respectively. The virtual screening of a similar collection of 370 of the compound 6KP and 139 of the compound A84 from PubChem revealed the compounds CID_66896495 and CID_11740546 as best inhibitors of MetAP II with an interaction energy equal to -40.27 KJ / mol for the first and -35.93 Kj / mol for the second. Finally, the Lipinski rule was performed to verify the pharmacokinetic properties of these two similar; they fit perfectly within the margin of the criteria imposed by Lipinski.
angiogenesis, molecular docking, FlexX, RMSD, methionine aminopéptidase II, Lipinski rule
Cancer is a real public health problem, figure among the main causes of morbidity and mortality in the world. The surgery, radiotherapy and chemotherapy are the three main ways to fight against this disease. Despite the progress, these methods kill both cancer cells and healthy cells, which induce many side effects. For this, new research to treat without inducing too many side effects [1]. As well, to grow and sustain its nutritional needs, the tumor will need to develop its own vascularization. The predominant mechanism to which it will resort is angiogenesis, which consists in the formation of new blood vessels from other, pre-existing [2].
In recent years, angiogenesis is a current track research for the treatment, and a new approach revolutionary promising uses the anti-angiogenic properties of certain molecules to try to block the cancer in depriving tumors of the nutrients and oxygen they need to grow in order to make them die of hunger. The endothelial cells, specialized in the development of new blood vessels, are the target of most of the anti-angiogenic strategies [3].
Anti-angiogenic targeted therapies act on the environment of the tumor; by inhibiting the formation of new blood vessels, they slow down tumor growth and the development of metastases. It is therefore necessary to identify new therapeutic targets for developing anti-angiogenic agents, and thus be able to propose treatments adapted to each patient [4].
In this context, the methionine aminopeptidase Type 2 (MetAP2) has an attractive potential therapeutic target interesting to develop new anti-angiogenic agents [5,6]. The MetAP is a metalloprotease that cleaves N-terminal methionine during protein synthesis, one of the critical steps in the maturation of the protein [7] and a member of a protein family that regulates the growth of endothelial cells.
By maintaining it at the molecular level, biology is based on the interaction between a protein and its substrate in most biological reactions. Understanding its mode of operation and defining the residues involved is therefore essential to explain the mechanisms that influence the affinity between two molecules.
In pharmacology, it is then possible to study these interactions by a large number of docking programs (commercial or free) that can objectively, rapidly and efficiently search for the relative orientation of a very large number of ligands within a target protein (virtual screening), which is considerably easier to implement, cheaper and faster than using an experimental method [8]. In silico molecular docking methods have already proved to be very successful and at present they have become crucial steps in the design of new bioactive molecules.
Therefore, the objective of this work is to:
• Demonstrate the best inhibitor of human MetAP2 from the interactions energies calculated by FlexX software in order to propose in silico new inhibitors more powerful towards the therapeutic target in question;
• Computer simulation of a collection of similar inhibitors selected in the previous step, in order to propose new inhibitors more powerful toward the MetAP2. These results will probably help develop an effective therapeutic tool to fight cancer development.
Preparation of molecules for docking
The complex MetAP-inhibitor is downloaded from the PDB database (http://www.rcsb.org) by introducing its ID code. The Flex X software requires the 3D structure for both molecules (protein - ligand). In addition, Flex X does not need a prior preparation; it uses directly the .pdb format for the enzyme and the .sdf format for the downloaded ligand.
FlexX Program
We have worked with the FlexX program (https://www.biosolveit.de) in the latest version 2.2.0. 2016, it was installed in our microcomputers.
FlexX is a commercialized computer software, currently part of LeadIT and is among the most used in molecular modeling, it allows to simulate the interactions between proteins and ligands, which consists of proposing new inhibitors and to assist the development of molecules with therapeutic activity.
Choice of a crystallographic structure
We chose a good quality code for the human MetAP2 enzyme 5JFR. The characteristics of this enzyme are shown in Table 1.
Table 1. Main characteristics of code 5JFR
Code |
Resolution (Å) |
Factor R |
Classification |
Number of chains |
Number of AA
per chain |
Number of atoms
per chain |
5JFR |
1.6 |
0.17 |
3.4.11.18 |
1 |
369 |
2897 |
Inhibition of 5JFR by Flex X
The choice of 5JFR: Among the 20 MetAP2-inhibitor complexes we chose the 5JFR code for our study (Figure 1), due to its good RMSD and its good resolution compared to the other complexes found in the PDB. In addition, the presence of a co-crystallized inhibitor.
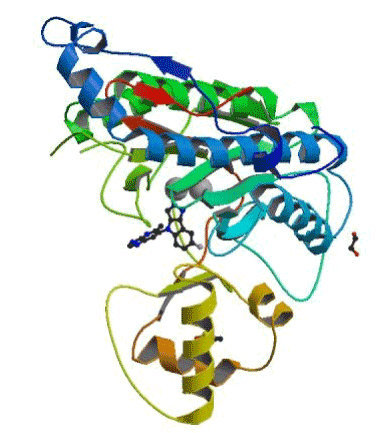
Figure 1. Structure of 5JFR.
Lipinski rule: Each possible drug must comply with several basic criteria, such as its low production cost, be soluble, stable, but must also comply with scales associated with its pharmacological properties of absorption, distribution, metabolism, Excretion and toxicity (ADME-T) [9]. This method is based essentially on Lipinski's rule 5 [10,11]:
- A maximum molecular weight of 500 g / mol;
- A maximum of 5 H-donor links;
- A partition coefficient (logP) ≤ 5;
- A maximum of 10 H-acceptor bonds;
- Number of rotary functions ≤ 15.
Calculation of interactions "enzyme- similar": To study the interactions between the MetAP2 enzyme and the similar we use a specific PubChem library.
PubChem: is an American database of chemical molecules managed by the National Center for Biotechnology Information (NCBI), a branch of the National Medical Library of the United States under the authority of the National Institutes of Health (NIH). PubChem currently lists several million compounds whose structure and physicochemical properties are accessible free of charge via the internet (http://pubchem.ncbi.nlm.nih.gov). This bank was used to obtain the structural analogues of our ligands 6KP and A84 to a percentage of similarity 90% in order to dock them by our software to search among these compounds one that binds more strongly than the 6KP and A84 to our target human MetAP2 and propose it as a new, more effective inhibitor. After having introduced the name of the chosen inhibitor, the properties of the latter will appear.
Inhibition of 5JFR by Various Inhibitors
We have selected four compounds that act on human MetAP2. The structures of the inhibitors studied are represented in Table 2.
Table 2. Structure of inhibitors studied
N° |
Inhibitor |
Code |
Name |
1 |
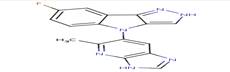
|
6KP |
7-fluoro-4-(5-methyl-3H-imidazo[4,5-b]pyridin- 6-yl)-2,4-dihydropyrazolo[4,3-b]indole |
2 |
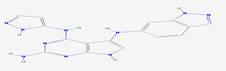
|
CID_66896495 |
5-N-(1H-indazol-6-yl)-4-N-(1H-pyrazol-5-yl)-7H-pyrrolo[2,3-d]pyrimidine-2,4,5-triamine |
3 |
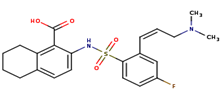
|
A84 |
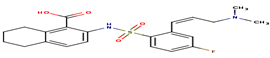
2-[({2-[(1z)-3-(dimethylamino)prop-1-enyl]-4 fluorophenyl}sulfonyl)amino]-5,6,7,8-tetrahydronaphthalene-1-carboxylic acid |
4 |
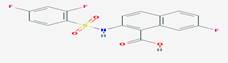
|
CID_11740546 |
2-[(2,4-difluorophenyl)sulfonylamino]-7-fluoronaphthalene-1-carboxylic
acid |
Study of the interactions involved in the inhibition of human MetAP2
Interaction 5JFR-Met
To study the mode of interaction of different inhibitors with the active site of the human MetAP2 enzyme by molecular docking; we used the latest version of the FlexX program, it shows the hydrophobic interactions and the hydrogen bonds, these last are the most interesting among the weak bonds.
The amino acids of the active site of the enzyme MetAP2: According to the FlexX program, the amino acids of the active site are: His231, Asp251, Asp262, Asn329, His331, His339, Glu364, His382, Tyr383, Asp442, Tyr444, and Glu459.
Our approach consists firstly of studying the interaction with methionine (Met), substrate of the target.
Figure 2 shows the active site of MetAP2 complexed with Met. The substrate is well centered in the active site of the enzyme. The ligand is represented as "lines" of different colors, and the amino acids of the active site are represented in "wireframe".
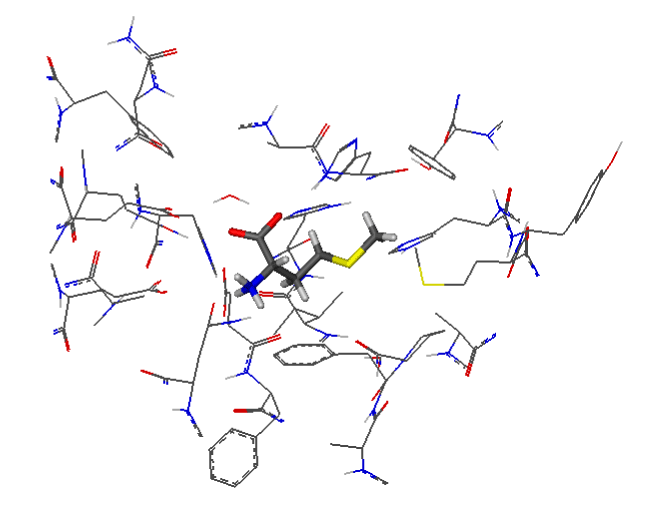
Figure 2. Docking of Met in the active site of 5JFR.
The visualization of the results of the docking by FlexX shows that the Met forms with the active site of the MetAP2 8 hydrogen interactions represented in discontinuous dashes:
-Two hydrogen bridges are observed on the one hand between the carbonyl of the Met and the NH group of the residue His231 and on the other hand with a molecule of water (O……..H-N His231) and (O…….H-O-H747);
-Two hydrogen bridges are observed between the carbonyl of the Met and a molecule of water (O……..H-O-H747) and (O…….H-O-H747);
- A hydrogen bond is observed between the carbonyl of the Met and the Zn2 + ion present in the active site (O……..Zn480);
- Two hydrogen bridges are observed between two carbonyl functions of the residue Asp262 with the NH group of the Met (N-H……..OAsp262) and (N-H…….. O Asp262);
- A hydrogen bond is observed between the amine function of the Met and the carbonyl of the residue Asp251 (N-H……..O Asp251).
There are hydrophobic interactions stabilizing the Met with the residues Ile338, Phe219 His231 and the ion Zn480 shown in green lines depicted in Figure 3.
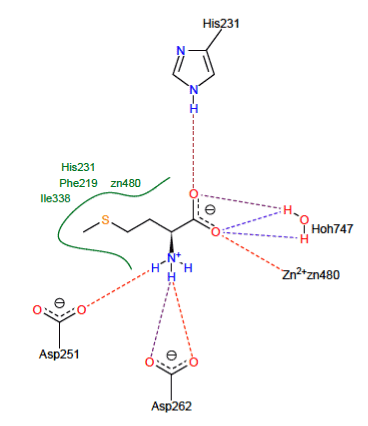
Figure 3. Representation of interaction formed with the substrate.
Interaction 5JFR-inhibitors
Docking : The docking of 20 molecules downloaded from the PDB with the .sdf format carrying different codes is carried out on the 5JFR crystallographic structure. We have considered interesting to test these inhibitors, and propose the best inhibitor of the enzyme MetAP2. The docking results are shown in Table 3.
Table 3. Docking results with the FlexX program
Number |
Inhibitor Code |
Score
Kj/mol |
Structure |
1 |
OVA |
-10.51 |
|
2 |
FUG |
0.00 |
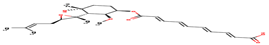
|
3 |
AO1 |
-22.17 |
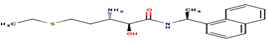
|
4 |
AO2 |
-25.02 |
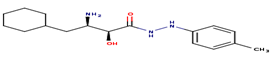
|
5 |
AO5 |
-24.43 |
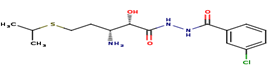
|
6 |
A41 |
-32.07 |
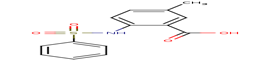
|
7 |
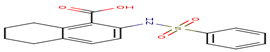
A75 |
-29.91 |
|
8 |
A84 |
-32.92 |
|
9 |
R20 |
-22.22 |
|
10 |
F77 |
-30.63 |
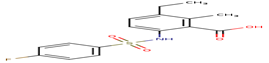
|
11 |
F79 |
-27,37 |
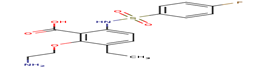
|
12 |
A19 |
-28,07 |
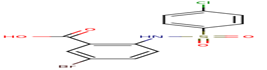
|
13 |
I96 |
-16,34 |
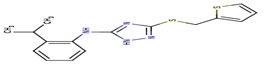
|
14 |
52P |
-20,59 |
|
15 |
94A |
-16,86 |
|
16 |
57R |
-15.2239 |
|
17 |
6KP |
-32,33 |
|
18 |
6KO |
-23,71 |
|
19 |
6KN |
-22,45 |
|
20 |
TN4 |
-10.17 |
|
It is useful to recall that among the 20 docked inhibitors, the compound 6KP (score = -32.33 Kj / mol) and compound A84 (score = -32.92 Kj / mol) with the best results (Table 4) present themselves as the Inhibitors the most powerful of the human MetAP2. For this we have chosen these compounds as a model in order to interpret their different interactions established with the enzyme MetAP2.
Table 4. Interaction energy of the best inhibitors
Ligand |
Score Kj/mol |
Match |
Lipo |
Ambig |
Clash |
Rot |
#match |
6KP |
-32.33 |
-27.64 |
-11.39 |
-5.96 |
7.27 |
0.00 |
25 |
A84 |
-32.92 |
-30.47 |
-11.81 |
-4.75 |
3.12 |
5.60 |
20 |
Lipinski Rule : Before starting the study of the interactions between the enzyme MetAP2 and the 2 inhibitors 6KP and A84; we applied the Lipinski rule (Table 5). This rule describes the physicochemical properties of a compound tested.
Table 5. Molecular properties of inhibitors studied
N |
Compounds |
PM (g/mol) |
nOH,NH |
nO,N |
ClogP |
Nrotb |
8 |
6KP |
306.304 |
2 |
4 |
2.7 |
1 |
17 |
A84 |
432.51 |
2 |
7 |
2.2 |
7 |
PM: Molecular weight; nOH,NH: Number of H-bond donors; nO,N: Number of H-bond acceptors; clogP: LogP or calculated partition coefficient; nrotb: Number of rotable links. |
The results of the table show that the two compounds 8 and 17 perfectly meet the criteria of the Lipinski rule. Both inhibitors are capable of exhibiting biological activity without having problems of oral absorption.
Visual analysis of the interactions "MetAP2 - inhibitors"
Interaction 5JFR-6KP : The visual analysis shows that the 6KP inhibitor is well placed in the active site of the enzyme MetAP2 (Figure 4). The complex is stabilized by the training of four hydrogen bonds:
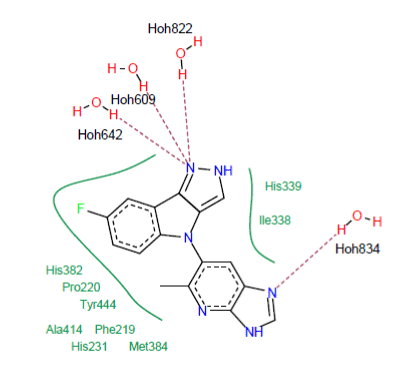
Figure 4. Mode of binding of the 6KP inhibitor to the active site of 5JFR
- A hydrogen bond is observed between the amine function of the 6KP inhibitor and a water molecule present in the active site (N……..H-O-H834);
- A hydrogen bond is observed between the amine function of the 6KP inhibitor and a water molecule present in the active site (N……..H-O-H822);
- A hydrogen bridge is formed between the amine function of the 6KP inhibitor and a water molecule present in the active site (N……..H-O-H609);
- A hydrogen bridge is formed between the amine function of the 6KP inhibitor and a water molecule present in the active site (N……..H-O-H642).
Several hydrophobic interactions stabilizing the 6KP inhibitor; they are carried out with the residues Ile338 His382, His339, Pro220, Phe219, Tyr444, Ala414, Met384, Ile338 and His231. The following figure visualizes these interactions:
Interaction : 5JFR-A84 : The FlexX program helped to view the number and type of links involved in the 5JFR-A84 interaction. The complex is stabilized by six hydrogen bonds (Figure 5):
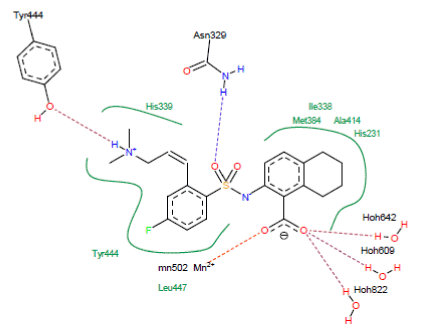
Figure 5. Method of binding inhibitor A84 with the active site of 5JFR
- A hydrogen bond is observed between the NH group of the inhibitor and the hydroxyl function of Tyr444 (N-H……..O-H-Tyr444);
- A hydrogen bond is observed between the carbonyl of the inhibitor A84 and the NH group of the residue Asn329 (O……..H-N-Asn329);
- Three hydrogen bonds between the carbonyl of the inhibitor A84 and the water molecules present in the active site (O….H-O-H642), (O….H-O-H609) and (O….H-O-H822);
- A hydrogen bridge between the carbonyl of the inhibitor A84 and the metal ion Mn2+ (O…….Mn2+).
Hydrophobic interactions stabilizing A84 with residues His339, Tyr444, Leu447, His231, Ala414, Ile338 and Met384.
Proposed new inhibitors of human MetAP2
Inhibition of the human MetAP2 by the similar of compound 6KP
In the aim of finding new more potent inhibitors of human MetAP2, we performed the molecular docking of a collection of 370 similar of the 6KP compound with 90% similarity. These chemical compounds are downloaded from the PubChem database. The results of docking by FlexX are presented in Table 6.
Table 6. The interaction energy values of the similar 6KP by FlexX
N |
Inhibitor code |
Score (Kj/mol) |
N |
Inhibitor code |
Score (Kj/mol) |
1 |
CID_60692209 |
-29.95 |
187 |
CID_91523782 |
-25.78 |
2 |
CID_60931616 |
-25.37 |
188 |
CID_90763912 |
-24.21 |
3 |
CID_65618114 |
-27.04 |
189 |
CID_90783106 |
-22.25 |
4 |
CID_65618518 |
-27.29 |
190 |
CID_91446673 |
-25.18 |
5 |
CID_66802281 |
-21.29 |
191 |
CID_76683822 |
-20.37 |
6 |
CID_66681148 |
-29.20 |
192 |
CID_73426341 |
-27.56 |
7 |
CID_66802416 |
-25.50 |
193 |
CID_89107435 |
-28.96 |
8 |
CID_66806570 |
-18.99 |
194 |
CID_73054826 |
-24.28 |
9 |
CID_66895607 |
-27.97 |
195 |
CID_71462984 |
-16.02 |
10 |
CID_66896495 |
-40.27 |
196 |
CID_71457622 |
-14.19 |
11 |
CID_67185223 |
-26.12 |
197 |
CID_73055705 |
-22.87 |
12 |
CID_67395425 |
-27.11 |
198 |
CID_71253882 |
-26.93 |
13 |
CID_67395981 |
-29.06 |
199 |
CID_71253683 |
-28.66 |
14 |
CID_67396135 |
-26.96 |
200 |
CID_71138132 |
-36.43 |
15 |
CID_67396548 |
-29.78 |
201 |
CID_70666195 |
-23.45 |
16 |
CID_67398150 |
-30.67 |
202 |
CID_69981294 |
-20.71 |
17 |
CID_67407477 |
-29.38 |
203 |
CID_69344609 |
-28.70 |
18 |
CID_67409220 |
-27.49 |
204 |
CID_69150294 |
-22.46 |
19 |
CID_67409331 |
-28.53 |
205 |
CID_68820816 |
-24.00 |
20 |
CID_67517105 |
-29.67 |
206 |
CID_69077400 |
-21.21 |
21 |
CID_67588149 |
-27.91 |
207 |
CID_69149290 |
-22.65 |
22 |
CID_67918391 |
-26.63 |
208 |
CID_69150316 |
-19.74 |
23 |
CID_68063317 |
-16.69 |
209 |
CID_69150551 |
-19.30 |
24 |
CID_68241781 |
-29.82 |
210 |
CID_66778322 |
-20.75 |
25 |
CID_68260700 |
-29.89 |
211 |
CID_66778621 |
-27.28 |
26 |
CID_68260703 |
-21.59 |
212 |
CID_66789083 |
-32.73 |
27 |
CID_68296446 |
-31.36 |
213 |
CID_66778918 |
-26.93 |
28 |
CID_67395425 |
-27.11 |
214 |
CID_67076481 |
-23.00 |
29 |
CID_68306301 |
-24.52 |
215 |
CID_66985044 |
-21.29 |
30 |
CID_68349946 |
-29.84 |
216 |
CID_67320203 |
-22.71 |
31 |
CID_68643830 |
-26.28 |
217 |
CID_68035521 |
-21.12 |
32 |
CID_68643860 |
-24.51 |
218 |
CID_59503887 |
-20.84 |
33 |
CID_68686238 |
-28.29 |
219 |
CID_59548207 |
-28.51 |
34 |
CID_69569483 |
-28.19 |
220 |
CID_59784965 |
-26.09 |
35 |
CID_69978572 |
-25.69 |
221 |
CID_59784880 |
-20.69 |
36 |
CID_70721644 |
-27.45 |
222 |
CID_59834985 |
-28.02 |
37 |
CID_70723293 |
-22.10 |
223 |
CID_60688217 |
-23.72 |
38 |
CID_70727168 |
-25.61 |
224 |
CID_66574664 |
-30.75 |
39 |
CID_70732987 |
-27.27 |
225 |
CID_66619888 |
-16.65 |
40 |
CID_70745872 |
-33.07 |
226 |
CID_59279329 |
-25.39 |
41 |
CID_70753996 |
-29.70 |
227 |
CID_59320844 |
-20.60 |
42 |
CID_70882179 |
-27.81 |
228 |
CID_59299145 |
-29.45 |
43 |
CID_70981612 |
-33.83 |
229 |
CID_59408914 |
-26.13 |
44 |
CID_71118180 |
-36.85 |
230 |
CID_59320938 |
-18.51 |
45 |
CID_71118184 |
-31.47 |
231 |
CID_59410548 |
-14.40 |
46 |
CID_123459036 |
-32.89 |
232 |
CID_59410552 |
-9.53 |
47 |
CID_123468906 |
-22.26 |
233 |
CID_59410557 |
-13.43 |
48 |
CID_71135894 |
-33.81 |
234 |
CID_58494345 |
-24.65 |
49 |
CID_71195295 |
-30.65 |
235 |
CID_58494361 |
-26.62 |
50 |
CID_71196481 |
-19.30 |
236 |
CID_58395561 |
-17.31 |
51 |
CID_71196595 |
-35.24 |
237 |
CID_58494377 |
-20.98 |
52 |
CID_71197129 |
-36.82 |
238 |
CID_58494431 |
-24.84 |
53 |
CID_71224460 |
-33.99 |
239 |
CID_58494433 |
-19.96 |
54 |
CID_71666619 |
-35.72 |
240 |
CID_59023024 |
0.00 |
55 |
CID_71721652 |
-24.80 |
241 |
CID_59053719 |
-24.13 |
56 |
CID_73333867 |
-33.05 |
242 |
CID_58335861 |
-24.36 |
57 |
CID_73425712 |
-26.53 |
243 |
CID_58335884 |
-26.20 |
58 |
CID_73425816 |
-26.50 |
244 |
CID_58382119 |
-21.48 |
59 |
CID_73426432 |
-32.13 |
245 |
CID_58394980 |
-17.80 |
60 |
CID_73426342 |
-19.97 |
246 |
CID_58395409 |
-22.00 |
61 |
CID_73426343 |
-27.12 |
247 |
CID_58395521 |
-22.32 |
62 |
CID_73426346 |
-33.34 |
248 |
CID_57917270 |
-21.09 |
63 |
CID_73426433 |
-29.44 |
249 |
CID_57803223 |
-28.83 |
64 |
CID_73426533 |
-25.84 |
250 |
CID_57917305 |
-23.05 |
65 |
CID_73426534 |
-23.67 |
251 |
CID_57917484 |
-22.27 |
66 |
CID_73426535 |
-24.88 |
252 |
CID_57990273 |
-31.92 |
67 |
CID_73426634 |
-22.53 |
253 |
CID_57917487 |
-21.92 |
68 |
CID_73426635 |
-23.33 |
254 |
CID_58212412 |
-22.07 |
69 |
CID_73426636 |
-16.61 |
255 |
CID_58164226 |
-21.09 |
70 |
CID_ 73426637 |
-24.64 |
256 |
CID_57803186 |
-25.50 |
71 |
CID_73426638 |
-21.77 |
257 |
CID_56971580 |
-26.38 |
72 |
CID_73426639 |
-24.65 |
258 |
CID_56971579 |
-23.36 |
73 |
CID_73426740 |
-15.55 |
259 |
CID_56971581 |
-21.67 |
74 |
CID_73426841 |
-19.98 |
360 |
CID_56971584 |
-24.57 |
75 |
CID_73426844 |
-26.58 |
261 |
CID_56971583 |
-24.69 |
76 |
CID_73426934 |
-25.84 |
262 |
CID_57477852 |
-20.78 |
77 |
CID_73426936 |
-25.53 |
263 |
CID_56911516 |
-26.63 |
78 |
CID_73426938 |
-22.62 |
264 |
CID_56914738 |
-25.91 |
79 |
CID_73427031 |
-27.55 |
265 |
CID_56964913 |
24.31 |
80 |
CID_73427128 |
-29.91 |
266 |
CID_56968277 |
-23.19 |
81 |
CID_73427131 |
-30.64 |
267 |
CID_56969870 |
-24.34 |
82 |
CID_73602825 |
-30.75 |
268 |
CID_56971364 |
-25.05 |
83 |
CID_73776964 |
-32.08 |
269 |
CID_53467204 |
-28.81 |
84 |
CID_73776965 |
-30.74 |
270 |
CID_56702964 |
-21.94 |
85 |
CID_73776966 |
-34.68 |
271 |
CID_56701575 |
-20.31 |
86 |
CID_73776969 |
-31.76 |
272 |
CID_56718837 |
-20.72 |
87 |
CID_73776982 |
-29.17 |
273 |
CID_56760588 |
-23.09 |
88 |
CID_73776983 |
-31.83 |
274 |
CID_56862202 |
-16.87 |
89 |
CID_73776985 |
-17.32 |
275 |
CID_56903702 |
-21.07 |
90 |
CID_73776986 |
-31.81 |
276 |
CID_56895085 |
-18.20 |
91 |
CID_73777001 |
-32.60 |
275 |
CID_56909010 |
-23.39 |
92 |
CID_73777002 |
-30.57 |
276 |
CID_56904736 |
-26.14 |
93 |
CID_73777003 |
-32.20 |
277 |
CID_56908944 |
-28.99 |
94 |
CID_73777041 |
-30.39 |
278 |
CID_53391408 |
-24.15 |
95 |
CID_73777093 |
-31.26 |
279 |
CID_53386318 |
-22.60 |
96 |
CID_73777094 |
-31.25 |
280 |
CID_53386266 |
-20.79 |
97 |
CID_73777095 |
-30.83 |
281 |
CID_53386265 |
-26.17 |
98 |
CID_73777096 |
-29.29 |
282 |
CID_53386212 |
-22.44 |
99 |
CID_73777097 |
-31.84 |
283 |
CID_53386262 |
-26.57 |
100 |
CID_73777117 |
-31.21 |
284 |
CID_53386065 |
-25.36 |
101 |
CID_73777120 |
-31.73 |
285 |
CID_53247380 |
-23.94 |
102 |
CID_73777141 |
-31.20 |
286 |
CID_53385608 |
-28.17 |
103 |
CID_73777142 |
-31.55 |
287 |
CID_53385610 |
-21.96 |
104 |
CID_73777143 |
-31.56 |
288 |
CID_53385694 |
-29.28 |
105 |
CID_73777160 |
-31.64 |
289 |
CID_52149343 |
-21.28 |
106 |
CID_73777180 |
-31.90 |
290 |
CID_52105283 |
-23.25 |
107 |
CID_73777182 |
-30.46 |
291 |
CID_52149466 |
-23.61 |
108 |
CID_73777203 |
-31.91 |
292 |
CID_52149467 |
-23.91 |
109 |
CID_73777204 |
-33.71 |
293 |
CID_52149468 |
-24.18 |
110 |
CID_73777206 |
-29.80 |
294 |
CID_52233282 |
-23.69 |
111 |
CID_73777207 |
-29.70 |
295 |
CID_50952457 |
-25.29 |
112 |
CID_73777231 |
-30.46 |
296 |
CID_50980221 |
-21.52 |
113 |
CID_73777229 |
-30.99 |
297 |
CID_52104075 |
-20.78 |
114 |
CID_73777233 |
-33.48 |
298 |
CID_52104076 |
-23.00 |
115 |
CID_73777254 |
-31.87 |
299 |
CID_89996909 |
-29.11 |
116 |
CID_73777255 |
-31.67 |
300 |
CID_89996980 |
-21.81 |
117 |
CID_73777256 |
-34.27 |
301 |
CID_89996985 |
-23.19 |
118 |
CID_73777257 |
-33.08 |
302 |
CID_89997015 |
-23.48 |
119 |
CID_73777276 |
-31.81 |
303 |
CID_89997016 |
-29.32 |
120 |
CID_73777277 |
-31.71 |
304 |
CID_89997020 |
-24.57 |
121 |
CID_73777278 |
-29.61 |
305 |
CID_89997025 |
-25.61 |
122 |
CID_73777279 |
-32.29 |
306 |
CID_89997096 |
-23.12 |
123 |
CID_75389929 |
-29.22 |
307 |
CID_90032792 |
-26.63 |
124 |
CID_76683823 |
-22.04 |
308 |
CID_90103748 |
-28.68 |
125 |
CID_76690840 |
-25.55 |
309 |
CID_90104859 |
-31.20 |
126 |
CID_77277816 |
-26.96 |
310 |
CID_90105333 |
-31.70 |
127 |
CID_78031119 |
-35.09 |
311 |
CID_90105351 |
-29.71 |
128 |
CID_78862342 |
-25.21 |
312 |
CID_90105393 |
-29.67 |
129 |
CID_85471419 |
-24.90 |
313 |
CID_90105584 |
-30.18 |
130 |
CID_85471421 |
-27.26 |
314 |
CID_90106563 |
-27.86 |
131 |
CID_85471819 |
-28.40 |
315 |
CID_90105777 |
-32.62 |
132 |
CID_87616463 |
-29.29 |
316 |
CID_90106134 |
-31.10 |
133 |
CID_87617017 |
-28.81 |
317 |
CID_90184906 |
-20.21 |
134 |
CID_89012982 |
-31.95 |
318 |
CID_90241411 |
-14.26 |
135 |
CID_89107436 |
-20.97 |
319 |
CID_90241443 |
-14.41 |
136 |
CID_89621494 |
-16.62 |
320 |
CID_90392446 |
-34.98 |
137 |
CID_89736646 |
-30.45 |
321 |
CID_90465087 |
-26.25 |
138 |
CID_89736652 |
-25.09 |
322 |
CID_95192271 |
-24.94 |
139 |
CID_89775969 |
-23.69 |
323 |
CID_95192272 |
-22.02 |
140 |
CID_89790507 |
-22.10 |
324 |
CID_95213677 |
-23.26 |
141 |
CID_89943353 |
-23.46 |
325 |
CID_95213678 |
-20.03 |
142 |
CID_89996896 |
-28.51 |
326 |
CID_95402502 |
-22.69 |
143 |
CID_95719490 |
-16.07 |
327 |
CID_95402503 |
-20.61 |
144 |
CID_95719491 |
-17.98 |
328 |
CID_123164286 |
-31.68 |
145 |
CID_97860688 |
-24.89 |
329 |
CID_123453257 |
-32.30 |
146 |
CID_97860689 |
-25.21 |
330 |
CID_101512968 |
-28.31 |
147 |
CID_6414464 |
-16,52 |
331 |
CID_16064939 |
-25,83 |
148 |
CID_6050889 |
-10,86 |
332 |
CID_15649470 |
-27,05 |
149 |
CID_6411753 |
0,00 |
333 |
CID_20286888 |
-12,97 |
150 |
CID_6817311 |
-10,58 |
334 |
CID_15136425 |
-27,12 |
151 |
CID_6800914 |
0,00 |
335 |
CID_20286932 |
-13,21 |
152 |
CID_6846696 |
-12,62 |
336 |
CID_20286895 |
-13,91 |
153 |
CID_10172014 |
-24,55 |
337 |
CID_20286979 |
-15,76 |
154 |
CID_9614625 |
-10,54 |
338 |
CID_20286892 |
-14,27 |
155 |
CID_10086264 |
-19,67 |
338 |
CID_20286983 |
-15,79 |
156 |
CID_10402605 |
-19,36 |
339 |
CID_20287054 |
-13,76 |
157 |
CID_11779677 |
-23,59 |
340 |
CID_20287064 |
-15,81 |
158 |
CID_10314503 |
-20,88 |
341 |
CID_20287021 |
-15,22 |
159 |
CID_15136426 |
-27,12 |
342 |
CID_22220154 |
-23,42 |
160 |
CID_22220163 |
-19,05 |
343 |
CID_4452867 |
-22,60 |
161 |
CID_24901328 |
-20,47 |
344 |
CID_44180844 |
-22,33 |
162 |
CID_24856686 |
-31,05 |
346 |
CID_43790603 |
-24,01 |
163 |
CID_22220135 |
-20,55 |
347 |
CID_44525865 |
-21,10 |
164 |
CID_25204149 |
0,00 |
348 |
CID_44333475 |
-25,37 |
165 |
CID_25204074 |
-7,66 |
349 |
CID_46239371 |
-34,11 |
166 |
CID_29152203 |
-25,63 |
350 |
CID_45236119 |
-18,38 |
167 |
CID_28735488 |
-23,84 |
351 |
CID_46174150 |
-24,14 |
168 |
CID_25204073 |
-13,62 |
352 |
CID_44622425 |
-21,99 |
169 |
CID_39893609 |
-18,32 |
353 |
CID_46238768 |
-27,88 |
170 |
CID_39893341 |
-16,97 |
354 |
CID_46239373 |
-26,17 |
171 |
CID_29153292 |
-20,79 |
355 |
CID_46239669 |
-25,41 |
172 |
CID_31105950 |
-22,80 |
356 |
CID_46239576 |
-34,93 |
173 |
CID_29258203 |
-23,64 |
357 |
CID_46239374 |
-29,03 |
174 |
CID_39796586 |
-32,20 |
358 |
CID_46994822 |
-24,93 |
175 |
CID_39893609 |
-18,32 |
359 |
CID_46985083 |
-19,81 |
176 |
CID_42196258 |
-16,97 |
360 |
CID_46994487 |
-24,87 |
177 |
CID_39893341 |
-16,97 |
361 |
CID_46992554 |
-16,01 |
178 |
CID_42196256 |
-18,23 |
362 |
CID_46987457 |
-18,55 |
179 |
CID_43790603 |
-24,01 |
363 |
CID_46997872 |
-22,99 |
180 |
CID_44333475 |
-25,37 |
364 |
CID_49784288 |
-26,27 |
181 |
CID_44180844 |
-22,33 |
365 |
CID_49785079 |
-28,44 |
182 |
CID_43782655 |
-23,67 |
366 |
CID_49784291 |
-26,31 |
183 |
CID_43788097 |
-26,36 |
367 |
CID_49785081 |
-30,28 |
184 |
CID_49785830 |
-25,55 |
368 |
CID_49765102 |
-25,65 |
185 |
CID_49825728 |
-27,31 |
370 |
CID_49784289 |
-26,18 |
According to the results of this table, the similar number 10 (CID_66896495) has a score (-40.27 Kj / mol), lower than that of the inhibitor 6KP (-32.33 Kj / mol).
From Table 7, it is found that the similar CID_66896495 has a molecular weight of lower than 500g/mol and a LogP of less than 5; which indicates a better lipophilic character. Therefore, we can say that this similar respond the rule of Lipinski. We considered it useful to detail the interactions involved with the active site of MetAP2.
Table 7. Criteria of the Lipinski rule for compound CID_66896495
N |
Compounds |
PM (g/mol) |
nOH,NH |
nO,N |
ClogP |
Nrotb |
10 |
CID_66896495 |
346.358 |
6 |
7 |
2.5 |
4 |
Visual analysis: The similar CID_66896495 forms with the active site of the MetAP2 seven hydrogen bonds:
- A hydrogen bridge is observed between the NH group of CID_66896495 and one of the ring nitrogen atoms of the His231 residue (N-H……..N His231);
- A hydrogen bond is formed between one of the nitrogen atoms of the ring of CID_66896495 and the NH group of the residue Asn329 (N……..H-N-Asn329);
- A hydrogen bond is observed between one of the nitrogen atoms of the ring of CID_66896495 and the NH group of the residue Asn329 (N……..H-N-Asn329);
- A hydrogen bridge is observed between the NH group of CID_66896495 and the carbonyl of the residue Asn329 (N-H……..O-Asn329);
- Three hydrogen bonds between one of the ring nitrogen atoms of CID_66896495 and the water molecules present in the active site (O….H-O-H642), (O….H-O-H609) and (O….H-O-H822).
In addition, this inhibitor establishes several hydrophobic interactions with the residues His339, Tyr444, His231 and Ile338 (Figure 6).
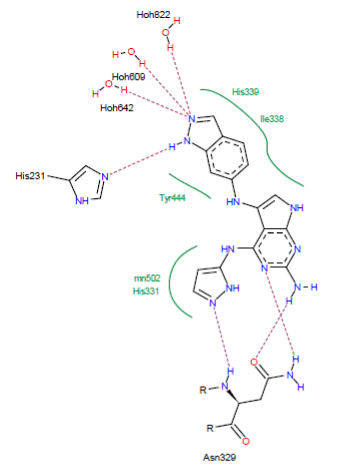
Figure 6. Representation of interactions formed by the similar CID_66896495
Inhibition of MetAP2 by similar to compound A84
Docking with FlexX, allowed us to calculate the energies of interaction of 139 similar of compound A84 from PubChem. These chemical compounds a percentage of similarity equal to 90%. The results are shown in the following table (Table 8).
Table 8. Docking results of the compounds of compound A84 with FlexX
N |
Inhibitor code |
Score(Kj/mol) |
N |
Inhibitor code |
Score(Kj/mol) |
1 |
CID_6102685 |
-19,85 |
71 |
CID_11597423 |
-19,23 |
2 |
CID_6102686 |
-20,01 |
72 |
CID_11597839 |
-18,12 |
3 |
CID_9997201 |
-19,13 |
73 |
CID_11604661 |
-16,22 |
4 |
CID_10067258 |
-23,55 |
74 |
CID_11627073 |
-17,81 |
5 |
CID_10043171 |
-22,24 |
75 |
CID_11613481 |
-21,56 |
6 |
CID_10023785 |
-23,50 |
76 |
CID_11633910 |
-17,62 |
7 |
CID_10246955 |
-21,38 |
77 |
CID_11633065 |
-17,75 |
8 |
CID_10316628 |
-22,08 |
78 |
CID_11597629 |
-14,01 |
9 |
CID_10427648 |
-22,42 |
79 |
CID_11633910 |
-17,62 |
10 |
CID_10452412 |
-23,66 |
80 |
CID_11634034 |
-18,28 |
11 |
CID_11503087 |
-19,09 |
81 |
CID_11641105 |
-20,50 |
12 |
CID_11503191 |
-14,68 |
82 |
CID_11670580 |
-15,00 |
13 |
CID_11510709 |
-13,49 |
83 |
CID_11634668 |
-14,02 |
14 |
CID_11510884 |
-16,36 |
84 |
CID_11691459 |
-14,07 |
15 |
CID_11511194 |
-16,24 |
85 |
CID_23644716 |
-20,64 |
16 |
CID_11517700 |
-19,11 |
86 |
CID_11723962 |
-21,47 |
17 |
CID_11517913 |
-21,30 |
87 |
CID_16102135 |
-20,36 |
18 |
CID_11533113 |
-16,87 |
88 |
CID_11719751 |
-17,32 |
19 |
CID_11540018 |
-18,54 |
89 |
CID_23647759 |
-19,55 |
20 |
CID_11546119 |
-14,97 |
90 |
CID_23647744 |
-15,89 |
21 |
CID_11546431 |
-20,56 |
91 |
CID_23647749 |
-13,56 |
22 |
CID_11554308 |
-15,28 |
92 |
CID_23647760 |
-20,41 |
23 |
CID_11561873 |
-17,74 |
93 |
CID_23647750 |
-21,93 |
24 |
CID_11568437 |
-18,69 |
94 |
CID_23647761 |
-20,25 |
25 |
CID_11569061 |
-18,86 |
95 |
CID_23647763 |
-19,32 |
26 |
CID_11569838 |
-22,18 |
96 |
CID_23647764 |
-18,00 |
27 |
CID_11573782 |
-20,84 |
97 |
CID_23647762 |
-18,95 |
28 |
CID_11576103 |
-18,97 |
98 |
CID_23647765 |
-17,28 |
29 |
CID_11576138 |
-17,39 |
99 |
CID_49823464 |
-18,00 |
30 |
CID_11576736 |
-20,93 |
100 |
CID_44413210 |
-21,46 |
31 |
CID_11577513 |
-21,14 |
101 |
CID_44413082 |
-23,82 |
32 |
CID_11583139 |
-20,51 |
102 |
CID_49823470 |
-19,48 |
33 |
CID_11583415 |
-19,47 |
103 |
CID_49823469 |
-17,35 |
34 |
CID_11583793 |
-19,55 |
104 |
CID_49823468 |
-18,80 |
35 |
CID_11590100 |
-21,99 |
105 |
CID_49823467 |
-23,85 |
36 |
CID_10472482 |
-34.38 |
106 |
CID_68863564 |
-32.77 |
37 |
CID_10473619 |
-34.29 |
107 |
CID_68863594 |
-21.80 |
38 |
CID_11494819 |
-30.58 |
108 |
CID_68863597 |
-21.22 |
39 |
CID_11496527 |
-20.10 |
109 |
CID_68863702 |
-31.19 |
40 |
CID_11518259 |
-17.25 |
110 |
CID_68863786 |
-16.88 |
41 |
CID_11518441 |
-22.92 |
111 |
CID_68863903 |
-30.44 |
42 |
CID_11539865 |
-26.92 |
112 |
CID_68863953 |
-30.62 |
43 |
CID_11540135 |
-24.92 |
113 |
CID_68863959 |
-30.60 |
44 |
CID_11561945 |
-21.49 |
114 |
CID_68864095 |
-29.53 |
45 |
CID_11577513 |
-31.13 |
115 |
CID_68864103 |
-29.69 |
46 |
CID_11641129 |
-28.21 |
116 |
CID_68864168 |
-34.45 |
47 |
CID_11650034 |
-24.92 |
117 |
CID_68864495 |
-29.94 |
48 |
CID_11688827 |
-30.93 |
118 |
CID_68864532 |
-18.73 |
49 |
CID_11705475 |
-30.82 |
119 |
CID_68864533 |
-18.73 |
50 |
CID_11725102 |
-35.80 |
120 |
CID_68864556 |
-27.68 |
51 |
CID_11726205 |
-35.81 |
121 |
CID_68864649 |
-18.01 |
52 |
CID_11740546 |
-35.93 |
122 |
CID_68864747 |
-29.90 |
53 |
CID_23647745 |
-28.40 |
123 |
CID_68864859 |
-18.31 |
54 |
CID_23647746 |
-28.37 |
124 |
CID_68865409 |
-32.57 |
55 |
CID_23647747 |
-28.90 |
125 |
CID_68865656 |
-30.90 |
56 |
CID_23647748 |
-29.33 |
126 |
CID_68865690 |
-30.94 |
57 |
CID_58534736 |
-25.69 |
127 |
CID_68866195 |
-29.72 |
58 |
CID_66692930 |
-28.06 |
128 |
CID_68866305 |
-30.14 |
59 |
CID_68862448 |
-20.40 |
129 |
CID_68866392 |
-29.78 |
60 |
CID_68862458 |
-18.01 |
130 |
CID_68866470 |
-26.86 |
61 |
CID_68862468 |
-30.32 |
131 |
CID_68866569 |
-30.62 |
62 |
CID_68862483 |
-25.57 |
132 |
CID_68866571 |
-30.42 |
63 |
CID_68862795 |
-30.70 |
133 |
CID_68978420 |
-31.39 |
64 |
CID_68863037 |
-25.46 |
134 |
CID_69705690 |
-31.43 |
65 |
CID_68863110 |
-27.73 |
135 |
CID_69708551 |
-31.48 |
66 |
CID_68963412 |
-29.15 |
136 |
CID_70640996 |
-25.84 |
67 |
CID_68863537 |
-35.42 |
137 |
CID_87605037 |
-28.27 |
68 |
CID_68863539 |
-19.20 |
138 |
CID_ 90971823 |
-28.78 |
69 |
CID_68863545 |
-33.48 |
139 |
CID_101846961 |
-29.90 |
70 |
CID_68863560 |
-30.59 |
|
|
|
2021 Copyright OAT. All rights reserv
According to the results of this table, the similar number 52 (CID_11740546) showed a score (-35.93 Kj / mol), lower than that of the inhibitor A84 (-32.92 KJ/mol). Table 9 shows that similar CID_ 11740546 studied perfectly meets the Lipinski rule.
Table 9. Criteria of the Lipinski rule for compound CID_11740546
N |
Compound |
PM |
nOH,NH |
nO,N |
ClogP |
Nrotb |
1 |
CID_11740546 |
381.325 |
2 |
8 |
4.1 |
4 |
Visual analysis : Visual analysis shows that the similar CID_ 11740546 forms five hydrogen bonds with the active site of the MetAP2.
- Two hydrogen bridges are formed on the one hand between the carbonyl of CID_11740546 and the metal ion Mn2+ and on the other hand with a molecule of water (O……Mn2+502) and (O…….H-O-H642) ;
- Three hydrogen bonds are formed between the carbonyl of CID_11740546 and the water molecules (O………...H-O-H609), (O………...H-O-H642) and (O………...H-O-H822).
The visualization of the docking results shows that similar CID_11740546 forms with the active site of the MetAP2 several hydrophobic interactions with the residues His231, Ala414, Tyr444, His331, Met384, Leu447, Leu328, Ile338, His382 and His339. Figure 7 shows these interactions.
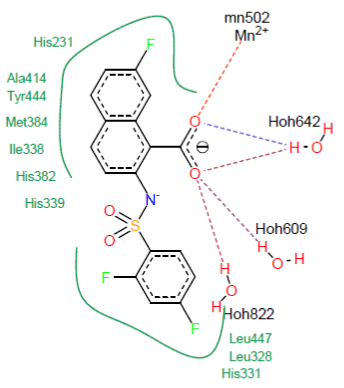
Figure 7. Representation of the interactions formed by the similar CID_11740546
The main objective of our study was to assist in the development of new inhibitors of methionine aminopeptidase II, a promising therapeutic target for the development of new anti-angiogenic drugs.
Our approach consisted in the first step of demonstrating the best inhibitor of MetAP2 among the 20 compounds derived from the PDB that we studied by FlexX. The two compounds 6KP and A84 being the best inhibitors, they have respectively the scores: -32.33 Kj / mol and -32.92 Kj / mol.
The second part was devoted to the search of new inhibitors of MetAP2. Computer simulation of a similar collection of 6KP and A84 inhibitors from PubChem; Allowed us to suggest compounds CID_66896495 and CID_11740546 as potential new inhibitors of MetAP2. Its energies of interaction less than those of 6KP and A84 and equal to -40.27 Kj / mol for the first and -35.93 Kj / mol for the second.
Finally, it is important to note that the two similar CID_66896495 and CID_11740546 tested by this study are consistent with the criteria imposed by the Lipinski rule, which are essential to allow the placing on the market of a possible drug.
To conclude, in the light of the results obtained in this work, which consists in the elucidation of the inhibition of the human MetAP2 by the molecular docking method in order to discover new anti-angiogenes, we propose the compounds CID_66896495 and CID_11740546 as potential new inhibitors of the enzyme.
- Roux A (2013) Cancer: une stratégie pour affamer les tumeurs. [En ligne]. Disponible sur: http://www.futura-sciences.com/sante/actualites/cancer-cancer-strategie-affamer-tumeurs
- Brauer C (2007) Les médiateurs et voies de signalisation intracellulaires impliqués dans le contrôle de l'angiogenèse tumorale: nouvelles stratégies thérapeutiques en cancérologie. Thèse de Doctorat en pharmacie. Nancy: Universite Henri Poincare. 179 P.
- Sancey L (2006) Évaluation d’un radioligand de l’intégrine αvβ3 (raft-rgd) Pour l’imagerie moléculaire de l’angiogenèse tumorale. Thèse de Doctorat en génie biologique et médical. Grenoble: Université joseph fourier. 254P.
- Schmitt C (2012) Expression, purification et cristallisation de l’Aminopeptidase-N humaine (APN ou CD13); évaluation in vitro et in vivo d’inhibiteurs sélectifs. Thèse de Doctorat en chimie biologie. Université de Haute-Alsace. 281P.
- Nonato MC, Widom J, Clardy J (2006) Human methionine aminopeptidase type 2 in complex with L- and D-methionine. Bioorg Med Chem Lett 16: 2580-2583.
- Boucherit H, Chikhi A, Bensegueni A, Merzoug A, Hioual KS, et al. (2013) Research of new molecules able to starve the tumors by molecular docking's method. Biochemistry & Pharmacology 2: 1-4.
- Olaleye O, Raghunand TR, Bhat S, He J, Tyagi S, et al. (2010) Methionine Aminopeptidases fromMycobacterium tuberculosis as Novel Antimycobacterial Targets. Chemistry & Biology 17: 86-97.
- Grosdidier A (2007) Conception d'un logiciel de docking et applications dans la recherche de nouvelles molécules actives. Thèse de doctorat en pharmacie. Grenoble: Université Joseph Fourier. France. 90 p.
- Miteva MA, Violas S, Montes M, Gomez D, Villoutreix BO (2006) FAF-Drugs: free ADME/tox filtering of compound collections. Nucleic Acids Research 34: W738–W744
- Lipinski CA, Lombardo F, Dominy BW, Feeny PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46: 3-26.
- Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S (2013) Cancer Incidence and Mortality Worldwide: IARC Cancer Base No. 11 Lyon, France: International
























