The author analyzed the intestinal microflora of patients with type 2 diabetes (T2DM) and investigated the relationship between the onset of T2DM and the intestinal microflora. The difference in fecal microbiota was not observed depending on the pathological degree of patients with T2DM pathology. There was a difference between fecal microbiota and microbiota in colon mucosal epithelium, but there was no difference to microbiota in each colon mucosal epithelium.
By the metagenomic analysis of fecal microbiota of patients with T2DM, it confirmed that microorganisms in volved in glycolysis, glucose metabolism-modified transferases, energy metabolism and glucose metabolism were present. In severe cases of patients with T2DM, microorganisms involved in the expression of the succinate dehydrogenase were observed. The succinate dehydrogenase is involved in the mitochondrial function of brown adipose tissue and activates energy metabolism. Patients with T2DM have mitochondrial dysfunction in brown adipose tissue, and the onset of T2DM is primarily due to mitochondrial dysfunction. It was suggested that gut microbiota induces microorganisms which promote energy metabolism in response to hyperglycemia and suppresses the rise in blood glucose.
There are many reports on the intestinal microflora and type 2 diabetes mellitus (T2DM), but the research results are different and the causal relationship has not been clarified [1-3]. It has not yet been clarified whether there is a difference in the intestinal microflora depending on the degree of T2DM symptoms. Furthermore, it has not been clarified microorganisms having what kind of physiological action are found in patients with T2DM. In this paper, the author investigated the above using techniques such as metagenomic analysis.
Metagenomic analysis of intestinal microflora in patients with T2DM, and the relationship intestinal microbiota and onset of T2DM.
Total eighty-eight patients with T2DM were recruited for this study. Fecal samples were collected from participants and genomic DNA was extracted and used for each analysis. The composition and diversity of the intestinal microflora was investigated in fecal DNA samples using Illumina sequencing of the V3-V4 region of 16sRNA. The analysis of microbiota in colon mucosal epithelium was performed in the same manner as feces.
A database for predictive analytics (KEGG) and PICRUSt software were used for the metagenomic analysis.
At the principal component analysis in fecal microflora, the difference did not show depending on the degree of pathology patients with T2DM. There was a difference between fecal microflora and microflora in colon mucosal epithelium, but there was no difference to microflora in each colon mucosal epithelium (Figure 1).
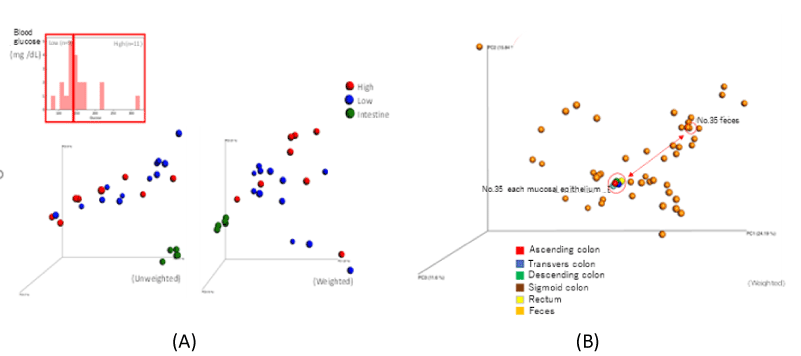
Figure 1. (A) PCoA analysis of fecal microbiota and microbiota of colon mucosal epithelium in patients with T2DM. (B) PCoA (Weighted) analysis of microbiota of colon mucosal epithelium and fecal microbiota in patients with T2DM
According to the metagenomic analysis in fecal microflora of patients with T2DM, it revealed that microorganisms involved in glycolysis [k16881 mannose-1-phophate guanylyl transferase], microorganisms involved in glucose metabolism-modified transferase [k03399 (gatD) cbi; cobalt-precorrin-7(C5)-methyltransferase], microorganisms involved in energy metabolism [k09482 glutamyl-tRNA (Gln) aminotransferase subunit D (gatD) cbiE; cobalt-precorrin-7(C5)-methyltransferase] and microorganisms involved in glucose metabolism [k00125 formate dehydrogenase] were present. In severe cases of T2DM, microorganisms involved in the expression of succinate dehydrogenase [k00242] were identified (Figure 2).
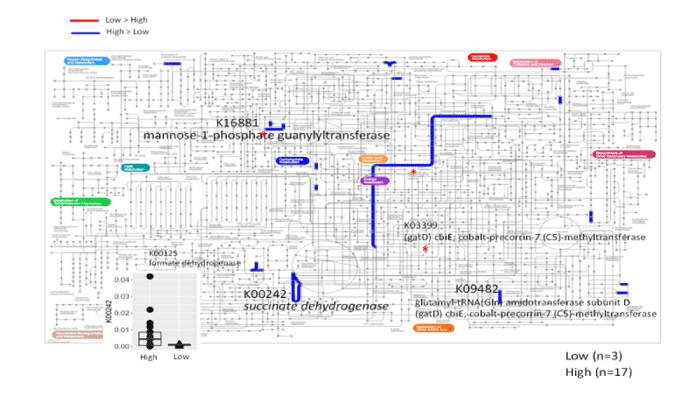
Figure 2. The metagenomic analysis of fecal microbiota in patients with T2DM
By the fecal metagenomic analysis of patients with T2DM, it confirmed that microorganisms involved in the below action existence.
・k16881 mannose-1-phophate guanylyl transferase: Involved in glycolysis
・k03399 (gatD) cbi; cobalt-precorrin-7(C5)-methyltransferase: Glucose metabolism-modified transferase involved in glucose metabolism
・k09482 glutamyl-tRNA (Gln) aminotransferase subunit D (gatD) cbiE; cobalt-precorrin-7(C5)-methyltransferase: Involved in energy metabolism
・k00125 formate dehydrogenase: In volved in glucose metabolism
・k00242 succinate dehydrogenase: Involved in the oxidation of succinic acid
The succinate dehydrogenase is involved in the mitochondrial function of brown adipose tissue and activates energy metabolism. Succinic acid enters brown adipose tissue mitochondria and increases energy cost [4]. In other words, when succinic acid accumulates in brown adipose tissue, succinic acid is converted to fumaric acid by succinate dehydrogenase by the electron transport chain of mitochondria in brown adipose tissue, and energy metabolism is enhanced. Mitochondrial function is divided into complex I to complex IV and complex V (Figure 3). In this way, the human body suppresses the rise in blood glucose level by increasing its own energy metabolism.
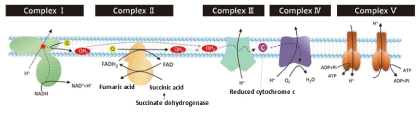
Figure 3. Mitochondrial electron transport chain system
The accumulated succinic acid is taken up by brown adipose tissue and rapidly metabolized by the mitochondria of brown adipose tissue to accelerate heat generation. Succinate metabolism requires its oxidation by succinate dehydrogenase.
It is known that patients with T2DM have mitochondrial dysfunction [5,6]. Therefore, the decisive factor for the onset of T2DM is considered to be mitochondrial dysfunction.
Intestinal microorganisms act to induce microorganisms involved in glycolysis, glucose metabolism, energy metabolism and expression of succinate dehydrogenase in response to the increase in blood glucose level. From the above, it was suggested that the intestinal microorganisms of patients with T2DM may work to maintain the homeostasis of the human body.
The intestine is the largest immune organ in the human body, and the intestinal flora that inhabits the intestine plays an important role in intestinal homeostasis, metabolism, and maintenance of immunity. This is also known as the “second genome” [7]. The gut microbiota plays an important role in the regulation of energy metabolism and inflammation and is closely associated with various chronic diseases such as obesity, T2DM, inflammatory bowel disease and rheumatoid arthritis [8-10]. In this analysis, it was shown that intestinal microorganisms may induce microorganisms which enhance energy metabolism when blood glucose rises to maintain homeostasis of the human body. Therefore, similar results should be obtained for also patients with IGT and T2DM reserve group as the pre-T2DM stage. The onset of T2DM is mainly due to mitochondrial dysfunction.
A method for screening the T2DM reserve group has not yet been developed. The development of the T2DM reserve screening method is the ultimate T2DM prophylaxis, and the development of the preliminary group screening method will have a great impact. The author believes that the fecal metagenomic analysis can develop a T2DM preliminary screening method. If the T2DM reserve group can be screened, treatments such as dietary guidance and fecal microbiota transplantation can completely prevent the onset of T2DM. The analysis of the intestinal microflora, which works to maintain the homeostasis of the human body, reflects the pathological condition earlier than the blood test, and is considered to be the main axis of preventive medicine in the future.
The fecal microflora of patients with T2DM were analyzed. There was no difference in fecal microflora depending on the degree of medical condition in patients with T2DM. In the metagenomic analysis of fecal microflora in patients with T2DM, a group of microorganisms involved in energy metabolism such as glycolysis, glucose metabolism and glucose metabolism modifying enzyme involved in glucose metabolism were observed. In severe cases of patients with T2DM, microorganisms involved in the expression of the succinate dehydrogenase were observed. The succinate dehydrogenase is involved in the mitochondrial function of brown adipose tissue and activates energy metabolism. Succinate dehydrogenase is involved in energy metabolism in brown adipose tissue mitochondria. Patients with T2DM have mitochondrial dysfunction in brown adipose tissue.
From these results, it was suggested that intestinal microflora may be involved in the maintenance of homeostasis of the human body.
There is no COI to disclose.
- Salgaço MK, Oliveira LGS, Costa GN, Bianchi F, Sivieri K (2019) Relationship between gut microbiota, probiotics, and type 2 diabetes mellitus. Appl Microbiol Biotechnol 103: 9229-9238.
- Vallianou NG, Stratigou T, Tsagarakis S (2019) Metformin and gut microbiota: their interactions and their impact on diabetes. Hormones (Athens) 18: 141-144.
- Tonucci LB, Dos Santos KMO, De Luces Fortes Ferreira CL, Ribeiro SMR, De Oliveira LL (2017) Gut microbiota and probiotics: Focus on diabetes mellitus. Crit Rev Food Sci Nutr 57: 2296-2309.
- Mills EL, Pierce KA, Jedrychowski MP, Garrity R, Winther S, et al. (2018) Accumulation of succinate controls activation of adipose tissue thermogenesis. Nature 560: 102-106.
- Pinti MV, Fink GK, Hathaway QA, Durr AJ, Kunovac A, et al. (2019) Mitochondrial dysfunction in type 2 diabetes mellitus: an organ-based analysis. Am J Physiol Endocrinol Metab 316: E268-E285.
- Kusminski CM, Ghaben AL, Morley TS, Samms RJ, Adams AC, et al. (2020) A novel model of diabetic complications: Adipocyte mitochondrial dysfunction triggers massive β-cell hyperplasia. Diabetes 69: 313-330.
- Lynch SV, Pedersen O (2016) The human intestinal microbiome in health and disease. N Engl J Med 375: 2369-2379. [Crossref]
- Festi D, Schiumerini R, Eusebi LH, Marasco G, Taddia M, et al. (2014) Gut microbiota and metabolic syndrome. World J Gastroenterol 20: 16079-94.
- Komaroff AL (2017) The microbiome and risk for obesity and diabetes. JAMA 317: 355-356. [Crossref]
- Cammarota G, Ianiro G, Cianci R, Bibbò S, Gasbarrini A, et al. (2015) The involvement of gut microbiota in inflammatory bowel disease pathogenesis: Potential for therapy. Pharmacology & Therapeutics 149: 191-212.



