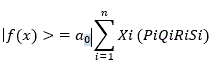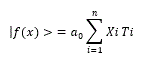Precision medicine requires for treatments according to the personalized genetic and environmental diversities. In insights of the diversity, individual variation of drug sensitivity would be implied in many factors, one of them would be the microRNA (miRNA) gene. The drug resistance would also be related with alteration of miRNA genotype in the cancer cell via its administrated anticancer drug. Further, natural products as the nutraceuticals could also modulate miRNA expression and promote anticancer effects. In the case of foods, not only foods’ miRNAs but also nutrients and natural products could render changing the phenotype of cells by alteration of miRNA expression. The facts show that environmental quantum energy of therapeutic drugs and foods may have an important role for miRNA gene expression upon the cells to personalize biological reaction under pharmacokinetics, and the new pharmacokinetics may be required through miRNA response. In turn, drugs among individuals treated would be also affected by personal state of quantum energy in the miRNA genes, and the quantum language of miRNA would dynamically adjust re-position of the personalized health. On the contrary, miRNA could control protein expression or epigenetic traits both negatively and positively, or directly and indirectly in the transcription and translation processes. Dysregulation of miRNA induces human cancer. Further, long noncoding RNA (lncRNA) and circular RNA (circRNA) could also act together with miRNAs and would control protein gene expression with miRNA. Participating in cancer development and progression, lncRNAs and circRNAs would directly or indirectly regulate signalling pathways and proliferation processes via miRNA sponging, and function as miRNA reservoir or as binding protein scavengers. The pharmaceutical agents would affect these RNA gene regulators at first in a cell and in vivo. To elucidate new pharmacokinetics of agents, the miRNA-mRNA-lncRNA-circRNA network architecture should be investigated. Thus, we review lncRNA and circRNA functions in cancers. Then, quantum energy relation among miRNA-mRNA-lncRNA-circRNA is discussed. Further, as a sample of drug treatment-related with quantum language of miRNA, drug resistance of human breast cancer was dynamically simulated using the miRNA entangling target sorter (METS). Since we have proved that quantum characters among miRNAs is implicated in oncogenesis, experimental evidences reviewed and dynamic in silico simulation with METS suggested that quantum language of miRNAs may be a common factor through tumorigenesis, anti-cancer and drug resistance.
microRNA, drug resistance, breast cancer, lncRNA, circRNA, food
While >90% of the human genome is transcribed, the dominant transcripts are noncoding RNA [1]. ncRNAs are classified as housekeeper and regulator, however, recently housekeepers of ribosome RNA (rRNA) and transfer RNA (tRNA) have been shown that they would produce miRNAs and transfer RNA fragments as the regulator [2,3]. Therefore, including long noncoding RNA (lncRNA) and circular RNA (circRNA), ncRNAs would be the regulators of protein expression [1,4-6]. miRNA is the noncoding gene while lncRNA is epigenetic regulator, but lncRNA also sometimes contains coding region or antisense of protein gene, further it remains forever so that the lncRNA is a source of the resident miRNA genes [7,8]. As described below, it seems to be doubtful what it is epi-gene because protein-coding DNA is absolutely transcribed to mRNAs and lncRNAs, which are also transcribed from protein-noncoding DNA, so it contains the protein gene. Thus, here, we define lncRNA and circRNA as two of the noncoding genes. These noncoding RNA (ncRNA) genes are implicated in treatment of chemotherapeutic agents, in diet as well as in immunotherapeutic agents. The new pharmacokinetic era has recently been revealed by miRNA-lcnRNA-circRNA network (Figure 1). In addition, while miRNA dysregulation in drug resistance of human breast cancer has been reviewed from several independent studies, using the miRNA entangling target sorter (METS), breast cancer drug resistance was, in silico, simulated for preparation of the personalized precision medicine [9].
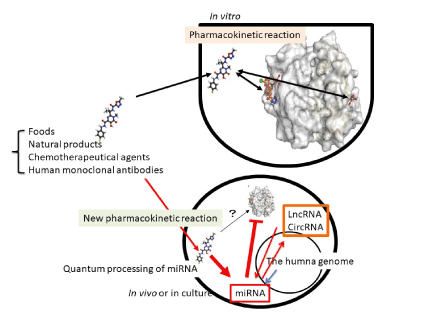
Figure 1. New pharmacokinetic pathway via ncRNAs. Theory of chemical reaction is based on equilibrium or non-equilibrium between substance A and substance B according to the law of mass action in vitro. However, ncRNAs, such as miRNA, lncRNA and circRNA are altered their expression pattern by not only chemical substances but also foods, monoclonal antibodies and ncRNA themselves in vivo. The quantum energy-bearing substances or quantum energy itself, such as temperature and electromagnetic wave would affect quantum language of ncRNAs in human cells. This new process should be involved into pharmacokinetics in vitro
Long noncoding RNA
LncRNA is also categorized as the regulator according to >200 nts long [8]. LncRNA class contains four types of the genomic organization (Figure 2).
Intergenic lncRNAs are derived from transcripts among protein genes. Intronic lncRNA is from introns. lncRNA overlapped exon and as antisense are the third and fourth classes. LncRNA acts as a transcriptional signal on one to one of Pol II, molecular decoys, guide of protein gene expression in cis and trans, and the scaffold molecule [4]. However, the four archetypes did not contain the relation to miRNAs, therefore, it has been explained that lncRNA, such as Air mapped in imprinting gene cluster lgf2r and would repress histone modification in allele-specific manner but in the imprinting site, methyltransferase or acetyltransferase expression could not be controlled by lncRNA. Although it has recently shown that lncRNA could function as a source of miRNAs bio-generated from primary RNA (pri-miRNA), a reservoir of miRNAs as the sponges, transcriptional activator and controller of pri-miRNA processing, etiology of cancer is deeply implicated in noncoding regulators of miRNA, lncRNA and circRNA [10-15]. For example, glioma is the lethal brain tumour. Hsa_circ_0046701 was expressed in glioma tissues, and KO of the circRNA 0046701 prevented glioma cell proliferation [16]. Silencing of circRNA 0046701 upregulated miR-142-3p and downregulated integrin subunit beta 8 (ITGB8). Glioma also upregulated lncRNA XIST and silencing of XIST suppressed metastasis and angiogenesis [17]. miR-429 inhibited XIST expression and suppressed glioma as a tumour suppressor. XIST also upregulated in endothelial cells of the blood-tumour barrier related with glioma. KO of XIST resulted downregulation of the transcription factor forehead box C1 (FOXC1) and zonula occludens 2 (ZO-2) by increasing expression of miR-137 [18]. PVT1 overexpression in glioma vascular endothelial cells induced protective autophagy by upregulation of Atg7 and Beclin I, which would be targeted by miR-186 [19]. PVT1 could be a sponge of miR-186. Further, HOXAas2 was increased in glioma cells [20]. HOXAas2 bound to miR-373 and miR-373 targets to epidermal growth factor receptor (EGFR). Since glioma malignancy is deeply implicated in vascularization process via EGFR signalling pathway, such as vascular endothelial-cadherin (V-cadherin), matrix metalloproteinase-2 (MMP-2) and -9 (MMP-9) through phosphatigylinositol 3 (PI3)-kinase pathway, HOXAas2/miR-373 interaction showed the relation of a molecular sponge. HOX transcript antisense RNA (HOTAIR) lncRNA expression was inversely related with miR-148b-3p in glioma cells, miR-141 as well [21-22].
MiR-34a/b/c has deeply been implicated in anti-tumour ability of p53 against wide spectrum of cancers [7]. LncRNA, LINC00473 was targeted by miR-34a in cervical cancer, lnc34c blocked miR-34a to bind miR-34a encoded gene in colon cancer stem cells [23,24]. About drug resistance of cancer cells, KO of lncRNA HOTAIR suppressed cisplatin resistance of gastric cancer by upregulation of miR-34a expression [25]. HOTAIR is a sponge of miR-34a. A soy isoflavone, genistein inhibited prostate cancer cells via reducing HOTAIR, resulting up-regulation of miR-34a [26]. It is suggested that natural product would be targeting to lncRNA and miRNA expression, probably through relation of quantum energy among chemical substances. In etiology of cancers, lncRNAs are implicated in cancers, such as breast cancer, however, the relation among miRNAs and lncRNAs has not been enough shown to simulate the network of miRNA/lncRNA/mRNA [27].
LncRNA would be indirectly regulated by protein expression through miRNAs. These data suggested that a lncRNA could be targeted by multiple miRNAs and a miRNA would be sponged by multiple lncRNAs. Those relations would be regulated through the similar rule to the implication between miRNAs and mRNAs. It means that mRNA as well as lncRNA would usually not be cleaved by the targeting of miRNAs and would functionally suppress to miRNA vice versa.
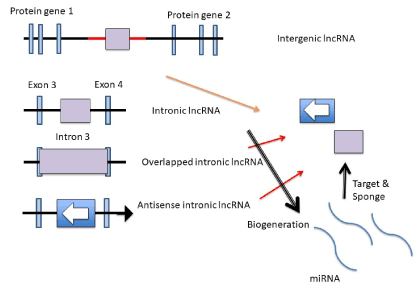
Figure 2.. LncRNAs’ bio-generation LncRNAs are produced from both intergenic and intronic DNAs in the human genome. Overlapped lncRNA contains protein coding region, and antisense lncRNA is the reverse direction of DNA sequences. The lncRNAs function as the target of miRNAs, sponge or resource of them.
Circular RNA (circRNA)
Circular RNA (circRNA) is belong to noncoding RNA (ncRNA) and is bio-generated from thousands of protein coding exon and intron (Figure 3).
The dominant circRNAs are cytoplasmic 3’→5’-linked ones consisting of exon (EcircRNA). The nuclear localized circRNAs are 2’→5’-linked ones consisting of intron (ciRNA). Spliceosome-dependent mechanisms lead to formation of circRNAs. Conventional collinear splicing causes the excision of intron from a multi-exon of protein gene and ciRNA is produced. Another nuclear localized circRNAs are 3’→5’-linked ones consisting of both exon and intron (ElciRNA). The 3’→5’-linked ElciRNAs are formed by back splicing. The back splicing is required for location of the 3’-end of an exon in closely proximity to an upstream 5’-end of the same or another exon. The looping structure by back splicing would be facilitated by the flanking reverse complimentary Alu retrotransposons or dimerizing RNA binding proteins. Further, circRNA could be bio-generated by an exon containing lariat from an exon-skipping events. The ciRNAs and ElciRNAs are secreted from cells and presented in blood, body fluids and tissues [28]. The function of circRNA against RNA gene information is implicated in the sponge effects to miRNAs and ciRNAs would be a source of intronic miRNAs as mitrons and simtrons [7]. Since LTR-retrotransposon, Alu and LINE have circular shape, and they have pre-miRNAs and miRNA target sequences, circRNA as well as the retroelements would facilitate the RNA crosstalk with miRNAs in the same functions. Since circRNA is bio-generated by splicing and the splicing is controlled by miRNAs, circRNA would simply have been a target of miRNA [29]. Although circRNA has other functions, such as stimulation of the initiation and elongation of RNA polymerase II transcripts by ElciRNA, RNA binding protein reservoir and translation of an exon including N (6)-methyl adenosine-driven translation of circRNA, these transcriptional and translational regulations would be restricted by canonical mRNA transcription and translation [28].
Therefore, circRNA and miRNA interaction would have more important role for protein gene expression to preciously control life events, such as cell proliferation and development than circRNA and protein direct interaction. In the competing endogenous RNA (ceRNA) theory, miRNAs could compete against target elements of mRNAs, long noncoding RNAs (lncRNAs) and circRNAs in the cytoplasm at the same time. If so, gain and loss effects of miRNA information could not be obtained from simple experiments on the bench. However, large amounts of research on the bench have showed that single miRNA gain and loss experiments affected the expression of target proteins. Thus, ceRNA is just the system but not language as information [30]. On the contrary, absolutely miRNA-miRNA quantum code would be information one [31].
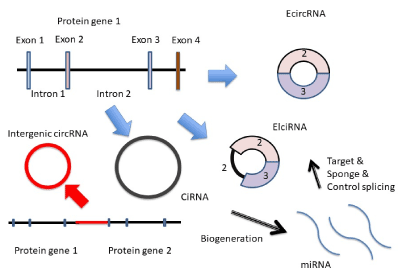
Figure 3. circRNAs’ bio-generation. circRNAs are produced from protein coding exon (EcircRNA), exon and intron (ElciRNA) and intron (CiRNA) or intergenic DNA. The circRNA works as miRNA target, sponge or resource of miRNAs.
Network computing analysis
To investigate the etiology of cancers from big data base using miRNA biomarkers, network analysis would be an important tool. The core networks and hubs of cancers have been reported by several network computing, such as glioma. Although the experimental data would have not been enough to complete molecular mechanisms of glioma, the network computing analysis revealed complex relation among miRNAs and target mRNAs [32]. About 34 altered miRNA expression was predicted in the glioma CpG island methylator phenotype (CIMP) (miR-19a/b, -1, -124, -206, -130a/b, -301, -29a/b/c, -494, -133a/b, -34/a/b/c, -449, -15a/b, -16, -186, -195, -424, -497, -140, -9, -302b, -145, -519a/b/c, -24, -506, and -122a). Further, glioblastoma has been investigated in lncRNA-mediated network prediction [33]. Chiu et al. has established the validation platform ‘Cupid’ for computation inferring network of miRNA-mRNA and lncRNA [34]. Cupid using machine learning prediction was trained on biochemical assay data, therefore, network analysis has been closed to be physiologically relevant at least. While retrotransposons are regulated by miRNA so that it is not sponge, expression of lncRNA is also controlled by miRNAs [35]. Meanwhile, miRNA target sites have also been proved themselves to locate in the protein coding RNA sequences, such as exon. Piwecka et al. have shown that knockout (KO) of Cdr1as/ciRS-7 circRNA in mouse brain causes depression of miR-7 and dysfunctional synaptic transmission [36]. Since miR-7 targets more than 70 binding sites of Cdr1as circRNA, exon, even noncoding and antisense, is actual target sites and miRNA did not break out the target of a sponge. Cdr1as would be degraded by miR-671 in Argonaute 2 (Ago2) dependent manner; however, Cdr1as was not cleaved by miR-7 [37]. KO of Cdr1as resulted decreasing of miR-7 but not increasing, whose results are clearly supported by mathematical model as following description. Further, knockdown of lncRNA ACT104 in osteosarcoma cells inhibited their proliferation, migration and invasion by upregulation of tumour suppressor miR-381 [38]. Thus, it is suggested that miRNA is still an axis of ncRNAs’ function because most of all proteins are directly controlled by miRNAs, and miRNAs would be tuned by other ncRNAs. Although we have been shown that HIV-1 N367 miRNA has the target site in various retroviruses and coding region has the target sites for N367, and rRNA-derived miRNAs have target sites in transposable element (TE) LINE, miRNAs would have physiochemically increased target sites according to ceRNA theory [2,30,39]. By simplified mathematical formula of quantum computing, it will be cleared. Tensor products of miRNA (X), mRNA (P), lncRNA (Q), circRNA (R) and TE (S) are shown in the qubits as previously described [31].
|ΨX > ⊗|ΨP> = |ΨXP>
|ΨX > ⊗|ΨQ> = |ΨXQ>
|ΨX > ⊗|ΨR> = |ΨXR>
|ΨX > ⊗|ΨS> = |ΨXS>
Tensor products of X, P, Q, R and S in a dimension are also presented as follows,
|ΨX > ⊗|ΨP> ⊗|ΨQ> ⊗|ΨR> ⊗|ΨS> = |Ψ >
Therefore, when i is 1, ----------, n,
|f(x)>, the total quantum qubit is linearly transformed with the vector spaces of Xi, Pi, Qi, Ri and Si.
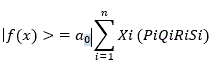
α0 is amplified value.
Xi, Pi, Qi, Ri and Si are the same formula as targets or sponges of miRNAs, therefore,
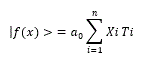
This formula is the same as that of DNSs among miRNAs [40]. From this formula, miRNA quantum energy can be accumulated, and it is strongly supported that plant or milk miRNAs are accumulated in transmitted organs [41-43] and plant miRNAs accumulate in human breast milk [44]. The cumulative effects have showed in quantum language simulation, such as miRNA entangling target sorting (METS) [45]. Since miRNA DNSs are corelated with CSCs, quantum energy between miRNA and mRNA depends upon DNS of miRNAs [46]. Although expression of the miRNA genes would genetically control upon the expression rates of mRNAs, lncRNAs, circRNAs and TE, miRNA expression could be controlled by miRNA itself in cancer [47]. By using the Cancer Genome Atlas (TCGA) data sets, the lncRNA-miRNA-mRNA network analyses have showed significant axis correlated with prognostic survival rate in lung squamous cell carcinoma [48]. The axis of network was PLAU mRNA, miR-31-5p, miR-455-3p, and lncRNAs, FAM83A-AS1, MIR31HG, and MIR99AHG. MIR31HG and MIR99AHG contain miR-31 and miR-99a host genes, respectively. Namely, since lncRNA, circRNA and TE are a source of pre-miRNAs and mature miRNAs, the increase-decrease rate would be involved in approximate parallel to that of miRNAs. Analyses of network among lncRNA, miRNA and mRNA have been performed in several cancers, such as pancreatic cancer, oesophageal cancer, hepatocellular carcinoma, colon cancer, rectal adenocarcinoma, gastric cancer, and thyroid carcinoma [49-55]. These data suggest that multiple miRNAs are the key driver to control mRNAs for cancer prognosis in the network.
Altogether, the resident miRNAs’ package would be stored with sponge ncRNAs and as reservoir ncRNA, and miRNA/miRNA interaction is more important for target selection than previous ceRNA [45]. Further, its sponge function shows the clear evidence that miRNAs are usually not degraded but the quantum energy of miRNAs are stored.
Pharmaceutical effects through ncRNAs
Chemotherapeutic agents react target proteins in vitro and it has been believed that such agents could work the same pharmaceutical activity in vivo as in vitro (Figure 1). Since first impact of agent transferred through the plasma membrane would be a chance meeting of large number of ncRNAs in the cytosol, miRNAs and lncRNA altered expression and tuned cell functions according to the response of ncRNAs against agent. Further, enzymes related with drug-metabolizing and transport, such as CYP450s and ABC or SLC transporters and xenobiotic receptors, could be controlled by the miRNA genes: therefore, if there were SNPs in miRNAs or in miRNA target sites, these genetic mutations are associated with chemotherapy response and clinical output [56]. It is suggesting that another pharmacokinetic era would be newly opened with the miRNA genes as well as their targets of lncRNA (Figure 1).
Natural products
Bufalin is a cardiotonic steroid from the Chinese medicine ChanSu. Treatment of Bufalin increased miR-203 expression and inhibited the stem cell-like phenotyping and induced apoptosis in glioma cells [57]. Bufalin prevented osteosarcoma cells through miR-148a [58]. Antipyretic analgesics in popular medicine, aspirin was correlated with expression of miR-145 in vascular smooth muscle cells, and its effects of anti-proliferation and anti-inflammation by aspirin were implicated in upregulation of miR-145 [59]. The omega-3 polyunsaturated acid (n-3 PFA), docosahexaenoic acid (DHA) reduced miR-21 oncomir in estrogen receptor-positive breast cancer cells and inhibited tumour growth [60]. The Chinese medical herb, tubeimoside-1 suppressed expression of miR-126-5p in non-small cell lung cancer cells and prevented proliferation of lung cancer cells via inhibition of vascular endothelial growth factor A (VEGF-A) [61]. Polyphenol, resveratrol derivatives piceatannol is well known as a strong antioxidant against cancer cells, but expression of miR-181a was increased by piceatannol treatment to melanoma cells [62]. Bcl-2 is a target of miR-181a, therefore, anti-cancer effect of piceatannol was implicated in upregulation of miR-181. Many reports showed that natural phytochemicals, resveratrol (3,5,4’-trihydroxy-trans-stilbene), epigallocatechin-3-gallate (EGCG), apigenin (4’,5,7-trihydroxyflavone), curcumin (diferuloylmethane) and honokiol induced alteration of miRNA expression in miR-663, miR-210, miR-122, miR-181b and miR-34a, respectively [63]. Although miRNAs modulate epigenetic enzymes, miR-203 by curcumin, miR-17-92 cluster by dietary butylate, diindolylmethane (DIM) by miR-34a, and miR-29a or miR-1256 by isoflavone would regulate epigenetic mechanisms for cancer suppression [64]. Further, ethanol enhanced miRNA and lncRNA transport via exosomes [65]. These results strongly suggest that natural agents would alter the miRNA gene expression, lncRNA and circRNA as well in vivo even if activities of chemical agents in vitro were distinct from miRNA functions [66]. The speculation has strongly been supported by in vivo clinical evidence that dietary supplements selenium and coenzyme Q10, known as an antioxidant, affected circulating miRNA expression in Swedish clinical trial [67].
Tamoxifen, human monoclonal antibody trastuzumab and doxorubicin
The relation between chemotherapeutic agents and miRNA has been reported in human breast cancer [68-70]. Approximate 70% of breast cancers have the estrogen receptor alpha (ERα), and 10-20% of them are the human epidermal growth factor receptor 2 (HER2) (ErbB2) positive. Triple-negative breast cancer accounts for 10-20%. Although ERα-positive tumour is treated with selective estrogen receptor modulators (SERMs), such as tamoxifen, endoxifen and toremifene, tamoxifen is predominantly treated for ERα-positive breast cancer because tamoxifen is a prodrug, which is bioactivated to endoxifen by CYP2D6 and CYP3A4 (Figure 4). Anti-HER2 human monoclonal antibody, trastuzumab is used for HER2 positive breast cancer. Triple-negative breast cancer did not have any target for its therapy, but doxorubicin would charge topoisomerase II and acts as DNA intercalator to inhibit DNA polymerization in breast cancer cells.
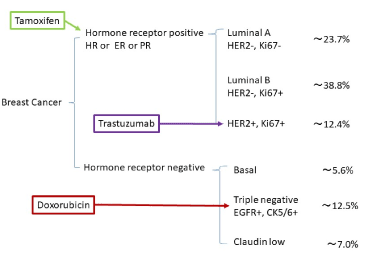
Figure 4. Human breast cancer subtype. Simple markers of human breast cancer are represented. Percentage of phenotypes are different between data of researchers; therefore, approximate percentages are shown. Tamoxifen, trastuzumab (Herceptin) and doxorubicin (Adriamycin) are administrated to ESR-positive, HER2-positive and triple negative breast cancer patients, respectively.
It is well known that ERα-positive, HER2-positive and triple negative breast cancer cells alter miRNA expression profile by treatment resistances of tamoxifen, trastuzumab and doxorubicin, respectively [71-76]. About etiological mechanisms from protein biology, tamoxifen resistant is implicated in phosphorylation of the serine 305 residue on ERα by protein kinase A (PKA) and p21-activated kinase-1. The 305-phosphorylation of ERα changes tamoxifen activity from antagonist of ERα to agonist but this mechanism is not fully cleared because endoxifen (tamoxifen) itself has no change binding to ERα and without chemical modification in culture [77]. In general, signal transduction pathways are positively related with gene expression but not negative regulation. Although AKT2-activated ERα (phosphorylation) contributes to tamoxifen resistance, PI3K binds together with ERα, and subsequently AKT2 and PI3K (PKC pathway far from PKA) activates ERα by a non-transcriptional and ligand-independent mechanism in vivo and in vitro [78]. But Sun et al. has not described about relation between PI3K/AKT2 and tamoxifen resistance of breast cancer [78]. ER α is also activated by growth factors, such as fibroblast growth factor 1 and 3 (FGFR 1 and 3) and insulin-like growth factor receptor 1 (IGF-1R). These growth factors are implicated in tamoxifen resistance via PI3K/AKT signalling [79,80]. The mechanisms of trastuzumab resistance are epitope asking, signalling alteration and immune response by monoclonal antibody Fcγ receptor polymorphism. However, mechanisms of tamoxifen, trastuzumab and doxorubicin resistance have not yet been understood at all. To further investigation of drug resistance, the role of ncRNAs have been reported in breast cancer. Trastuzumab-responsible miRNAs were observed in trastuzumab-resistant breast cancer patients (HER-2 positive breast cancer) [81]. miR-210 was upregulated in trastuzumab-resistant BT474 cells in vitro. On the other hand, trastuzumab treatment of BT474 decreased miR-194 expression [82]. In the culture of trastuzumab resistant breast cancer cell, SKBr-3, miR-200c was significantly reduced [83]. Further, miR-129-5p was downregulated in trastuzumab-resistant human breast cancer cells (HER-2 positive breast cancer), JIMT-1 in vitro and in sera of patients [84]. Overexpression of miR-129-5p increased sensitivity to trastuzumab in JIMT-1. In the case of HER-2 positive gastric cancer, upregulation of miR-125b was significantly associated with trastuzumab resistance of HER-2 positive gastric cancer and was implicated in malignant progression and poor prognosis [85]. These data suggest that protein agents could affect expression of miRNAs in treated cancer cells and antibody agent has new pharmacokinetic activity to modulate miRNA expression. LncRNA ATB was upregulated in trastuzumab resistant SKBr-3 cells, and lncRNA GAS5 was downregulated in the same cell line and tissues from trastuzumab-treated patients [86,87]. The former was the sponge of miR-200c and the latter was that of miR-21. Chemotherapeutic anti-EGFR, lapatinib upregulated GAS5 in trastuzumab resistant SKBr-3, suggesting that functionally different agents show pharmaceutically distinct effects against lncRNAs [87]. On the contrary, miR-21 increasing was correlated with treatment of both trastuzumab and anthhracycline- or taxane-based chemotherapy in HER-2 positive breast cancer [88]. Therefore, responses of lncRNA GAS5 and miR-21 were different against each treatment in breast cancer at all. Further, since miR-21 would work as a gene but lncRNA would be a target, in actual, two RNAs would have been given upon the distinct function because the proof of concept for the miRNA gene has recently been finally confirmed in conserved miRNA loci of the inbred mouse strains’ genome [89]. Although as described above, miRNA-miRNA interaction is important, Cilek et al. have simulated in the network analysis that trastuzumab-responsible miRNA-miRNA network has strong responsible for drug response in SKBr3 and BT474 breast cancer cells [90]. They found that miR-3064-3p and miR-32-3p were deeply connected to miR-216b and these miRNAs target YWHAE, RPL37 and AK2, which are related with apoptosis, cell cycle and metabolic pathway. Although alteration of miRNAs has been reported in trastuzumab resistance, trastuzumab treatment alters miRNA expression in HER2-positive breast cancer and would result in the resistant state [91]. We selected miRNAs for the MMP of trastuzumab resistance as downregulation of miR-200c, miR-221 and miR-205-5p, and upregulation of miR-375, miR-7-5p, miR-542-3p, miR-21, miR-210 and miR-515 (Table 1). Finally, pathway of antibody-receptor was regulated by miRNAs, therefore, drug affects expression of miRNAs directly and indirectly that is far from in vitro pharmacokinetic data, and the activity of drug in vivo cannot be correctly measured in vitro. Targets of trastuzumab resistance are mainly EGFR or PIK3/AKT in miRNA researches (Table 1), which shows an agreement with previous description about protein biology.
Table 1. Trastuzumab resistant related miRNAs in breast cancer
|
Drug
|
miRNA
|
Target
|
Mechanism
|
SNS
|
Reference
|
|
Trastuzumab
|
**miR-375
|
IGFIR, AKT
|
Activate P13K/Akt pathway
|
7
|
104
|
|
**miR-7-5p
|
EGFR
|
Enhance EGFR expression
|
7
|
105
|
|
*miR-200c
|
ZNF17
|
Promote TGF-beta signalling
|
6
|
83
|
|
**miR-542-3p
|
AKT
|
Activate P13K/Akt pathway
|
5
|
125
|
|
**miR-21
|
PTEN, PDCD4
|
Activate P13K/Akt pathway
|
5
|
126
|
|
*miR-221
|
PTEN
|
Activate P13K/Akt pathway
|
4
|
73
|
|
**miR-210
|
MET, IGF1R
|
Tumur growth
|
4
|
81
|
|
*miR-205-5p
|
P63
|
Promote EGFR pathway
|
4
|
127
|
|
**miR-515
|
MARK4
|
Supression PIK3C/MARK4
|
3
|
128
|
*: downregulation; **: upregulation
Simulation models of tamoxifen, trastuzumab and doxorubicin resistances
To elucidate the pathophysiological interactions between miRNAs and human breast cancer for personalized medicine, etiological causes of drug resistance were investigated by dynamic computer simulation in human breast cancer and its resistance according to systemic treatment. Since profiles of miRNAs have been available for prediction of human diseases, the microRNA memory packages (MMPs) were applied for the prediction of difference in drug resistance [46]. About in vivo trastuzumab resistance in breast cancer, 7 miRNAs were used, and it was compared with doxorubicin and tamoxifen resistances (Figure 5), in another word, at first contents of drug resistance were simulated before the context.
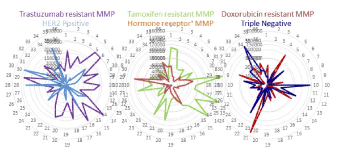
Figure 5. MMPs related with human breast cancer. Six MMPs of HER2-positive, ESR-positive, triple negative, trastuzumab resistance, tamoxifen resistance and doxorubicin resistance were computed and shown as radar chart.
Back bones of drug resistant MMPs are ERα positive, HER2 positive and triple negative breast cancer ones. All MMPs were showed as unique shapes, and 6 types (three inputs and three outputs) of MMPs were obtained (Figure 5). Although human breast cancer is a heterogeneous disease, it seems that drug resistance would also have a heterogeneous phenotype. However, it is possible to be diagnosed by miRNA comb about prediction of drug resistance of human breast cancer, and it is strongly supported by which each MMP is unique. As artificial intelligence (AI) is programmed by deep learning algorithm into the layer architecture (Figure S1), therefore, we firstly screened drug resistant-related miRNAs in both downregulation (Figure S2 blue characters) and upregulation (Figure S2 red characters) to pick up from the PubMed and Google Scholar databases.
From matrix of DNSs in SNSs of selected miRNAs, layer structures were constructed depending on the quantum energy levels (Figure S3) and depicted as mountain views (Figure S4). Three-dimension (3D) map was represented according to 10 DNS contour lines about DNSs of trastuzumab, doxorubicin and tamoxifen resistant-related miRNAs.
Next, context of drug resistance was simulated to understand difference of MMP. Since the network related with cancer has been dynamically computed by microRNA entangling target sorting (METS) algorithm, the network was predicted by METS simulator about in vivo ERα positive in breast cancer and it was compared with the network of tamoxifen resistance [45]. DNSs of 9 miRNAs in the matrix of tamoxifen resistance were calculated and top 4 values of DNS (70-80 called as quantum code region, QCR) were selected as miR-375, miR-342-5p, miR-320a and miR-378a-3p. The multi-targets of four miRNAs were searched in miRTarBase V21 (mirtarbase.mbc.nctu.edu.tw/php/index.php) and TargetScan 7.2 (www.targetscan.org/vert_72/) in human, then, target genes and related miRNAs were simulated by METS. The multi-miRNAs in a protein mRNA target were also reversely selected according to the top 10 DNS. Top layer (QCR: 70-80) of tamoxifen-resistant-related miRNAs were compared with the same layer (QCR: 70-80) of ERα positive breast cancer in target protein-protein interaction and the cluster on String (https://string-db.org/cgi/input.pl) (Figure 6A). Finally, protein gene function was searched by GeneCards (http://www.genecards.org) and validation of statistical significance in the simulation, such as the area under the ROC curve (AUC) or the χ2-based Cochran’s Q test was computed by BellCurve for Excel (Social Survey Research Information Co. Ltd., Tokyo, Japan). About searching of lncRNA, Long Noncoding RNA Database was used (www.lncrnadb.org). In ERα positive breast cancer (Figure 6a), three core miRNAs, let-7a-5p, miR-34a-5p and miR-206 are downregulated. Under METS simulation (Figure 6b), ERα would be upregulated by miR-206 and miR-18a-5p (miR-17-92 cluster) downregulation as tumour suppressors of breast cancer [92-94]. Downregulation of miR-34a-5p contributes suppression of TP53 in breast cancer [95,96]. Decreasing let-7a (let-7 family) and miR-34a is correlated with anti-apoptotic pathway [97]. After resistance of tamoxifen treatment, miR-375 downregulation would increase PI3K expression (Figure 6c), which has induced activation of ERα together with AKT as described above. As shown in GO enrichment analysis (geneontology.org/page/go-enrichment-analysis) (Figure 6c), cytoplasmic region would be a component of tamoxifen resistance under miR-375 targets, such as PI3K. On the contrary, targets of miR-206 and let-7 family were changed from ERα, and apoptosis-related transcriptional regulator, high mobility group A1 (HMGA1) to cytoplasmic vesicle-related complexin 2 (CPLX2) and chaperon protein, translocase of inner mitochondrial membrane (TIMM), respectively. These simulations strongly supported that alteration of ERα phosphorylation from PKA to PKC pathways is dominantly implicated in tamoxifen resistance in human breast cancer.
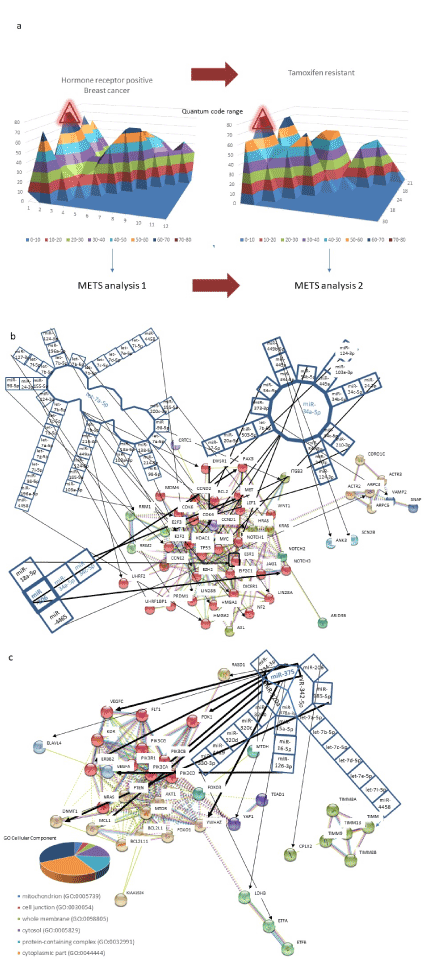
Figure 6. METS simulation of tamoxifen resistance in human breast cancer. ESR-positive human breast cancer (METS analysis 1) was treated with tamoxifen and then cells obtained a drug resistance as the acquired character (METS analysis 2) (a). From quantum energy level diagram, top level of layers (QCR: 70-80) had compared each other. METS analysis 1. Three miRNAs, miR-34a-5p, let-7a-5p and miR-206 in 70-80 QCR were selected in the simulation of ESR-positive breast cancer (b). Then, target protein genes were presented as the network among miRNAs. METS analysis 2. Four miRNAs, miR-342-5p, miR-375, miR-320a and miR-378a-3p in 70-80 QCR were analyzed in the next simulation of tamoxifen resistance to be compared with METS analysis 1 (c). Target protein genes were also shown. The clustered genes (red circles) were further characterized by GO cellular component enrichment.
In the case of HER2 positive breast cancer, the networks of QCR 90-100 and QCR 40-50 were investigated at first, and then those of HER2 positive and trastuzumab resistance were compared at the same QCR-levelled layers (QCR 40-50) by METS (Figure 7a). As shown in Figure 7b, downregulation of miR-134-5p, miR-637 and miR-193a-5p would induce upregulation of STAT, nuclear factor I/C (NFIC) and ribosomal protein S2 (RPS2). STAT and NFIC are transcriptional activator via HER2 signalling and RPS2 is a component of the 40S subunit, respectively. RPS2 is associated with Diamond-Blackfan Anaemia as ribosomopathy and activates mRNA production via formation of the cap-binding complex and eIFs [2]. Further, these three mRNAs and their comb miRNAs would cooperate with regulation of mTOR, PI3K and KRAS. The HER is belong to the epidermal factor receptor (EGFR) tyrosine kinase receptor family and amplification of it stimulates MAPK, PI3K and mTOR pathways [98]. This evidence is equivalent to our prediction by METS.
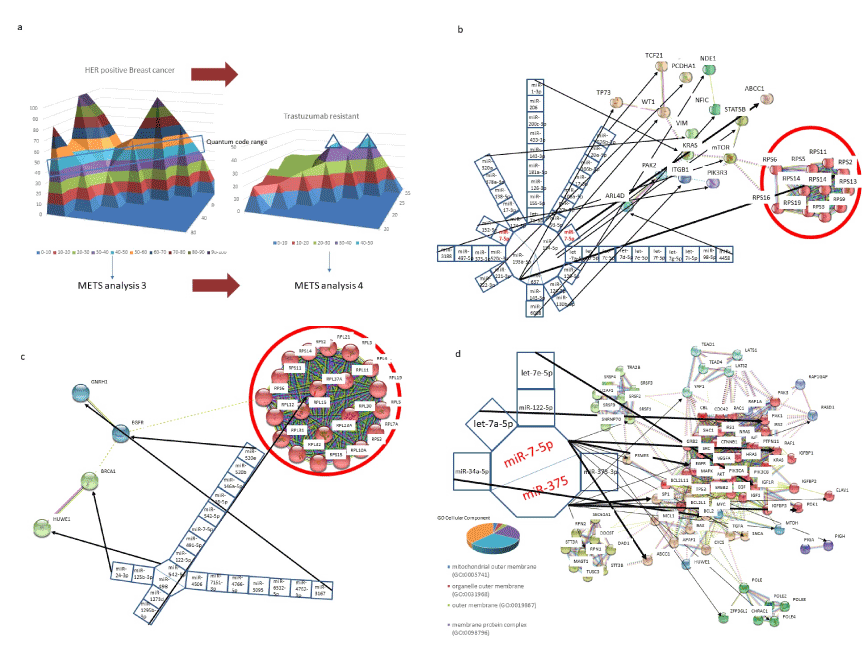
Figure 7. METS simulation of trastuzumab resistance in human breast cancer. HER2- positive human breast cancer (METS analysis 3) was administered with trastuzumab (a), and cells obtained a drug resistance (METS analysis 4). The QCR 40-50 was compared with each other. METS analysis 3.1. Three miRNAs, miR-134-5p, miR-193a-5p and miR-637 in 90-100 QCR were selected in simulation of HER2-positive breast cancer in advance (b). A large red circle contains robosomopathy-related protein gene cluster. METS analysis 3.2 (c). To compute the network on trastuzumab resistant breast cancer, at 40-50 QCR two miRNAs, miR-542-5p and miR-498 were selected and computed by METS. A large red circle also contains robosomopathy-related protein gene cluster. METS analysis 4 (d). At QCR 40-50, two miRNAs, miR-7-5p and miR-375 were selected in trastuzumab resistant breast cancer. The clustered genes (red circles) were further characterized by GO cellular component enrichment.
In the layer of QCR 40-50 (Figure 7c), downregulation of miR-498, human specific miR-1273d and miR-1295b-3p are linked in RPL37A, which is a component of 60S subunit of ribosomal complex. Therefore, HER2 positive human breast cancer would be involved into a case of ribosomopathy and this results from simulation would be a new insight of etiology in HER2 positive human breast cancer. Human breast cancer has been implicated in ribosomopathy, however, etiological causes of dysregulation of ribosomal proteins have not yet been explained with their gene- or oncoprotein gene- and tumour suppressor protein gene-, amplification, deletion, mutation and SNPs in the data from TCGA [99]. Thus, it is suggested that dysregulation of miRNAs could induce ribosomopathy, and anti-ribosomopathy would target development of agents against human HER2-positive breast cancer. Furthermore, miR-498 and its comb miRNAs are implicated in tumour suppressor, BCCA1, and miR-542-5p would target EGFR. After treatment of trastuzumab, most of patients respond trastuzumab resistance within a year [100]. As shown in Figure 7d, miR-375 can target SP1 and miR-7-5p targets ABCC1. MicroRNA package of miR-375 and miR-34a-5p targets BCL2 and that of miR-7-5p, miR-122-5p targets IGF1R. ABCC1, BCL2, IGF1R and SP1 are connected as cell membrane signalling and tumour proliferation. To validate the prediction by METS, dominant relations (red circles) were applied for GO cellular component analyses (Figure 7d). It is predicted that miRNA-miRNA interactions of trastuzumab resistance would be implicated in cell membrane signalling from these GOs. Five of the seven miRNAs (AUC; 0.71, p<0.01) have been related with breast cancer [101-103]. Since miR-7-5p and miR-375 are tumour suppressor of breast cancer and both are implicated in trastuzumab resistance of HER2 positive breast cancer, especially, miR-7-5p expression sensitized HER2∆16 to trastuzumab, predicted proteins would be therapeutic targets of HER2-positive breast cancer resistant to trastuzumab [104,105]. In the case of trastuzumab resistance, total target prediction data by METS statistically shows the area under the curve (AUC) 0.90 (P<0.001) in receiver operating characteristic (ROC). Thus, it is suggested that humoral and cellular immune reactions via human monoclonal antibody is controlled by miRNA genes.
Layers of quantum code in miRNAs were investigated in the triple negative breast cancer (Figure 8a). QCR 40-50 in triple negative breast cancer was compared with that in doxorubicin resistant breast cancer. In addition, top layer of doxorubicin was also simulated by METS. Although triple negative breast cancer is characterized by no achievement of complete response with chemotherapy, prediction of chemotherapy response needs to be addressed. In QCR 40-50, upregulation of miR-375-5p would inhibit activation of tumour suppressors, PTEN and a Wnt signalling component, CTNNB1 (Figure 8b). Further, downregulation of miR-200a-5p would increase TP53 suppressor, MDM4. Although amplification of 8p11-12, including LSM1, BAG4 and C8orf4 was observed in approximate 15% of breast cancer, activation of LSM protein component of LSM6, which is involved in mRNA degradation, was also simulated in QCR 40-50. On the other hand, doxorubicin resistant breast cancer upregulated miR-181a-5p and downregulated miR-106a-5p (Figure 8c) [106]. By treatment of doxorubicin, upregulation of miR-181a-5p would have still suppressed expression of anti-apoptotic protein BCL2 and downregulation of miR-106a-5p would have enhanced RB1 tumour suppressor even if in drug resistant cells. Therefore, doxorubicin resistant cancer cells would be likely to result drug sensitive in an anti-cancer state in QCR 40-50; in the top layer of QCR 90-100, however, downregulation of miR-663a would control AP-1 complex in GO cellular component analysis (Figure 8d, red circles genes). In addition, lncRNA H19/miR-675-5p axis is oncogenic [107]. It is suggested that high layer would administer the low layer, therefore, top layer-targeting agents would be necessary for therapy against doxorubicin resistant breast cancer.
Altogether, drug-resistant phenotypes of breast cancer were classified as 1) the substitution of main players in miRNAs on the same quantum energy level, 2) alteration from the hierarchy longitudinal carcinogenic mechanism, such as ribosomopathy, to the quantum level specific tumorigenic one 3) the higher quantum level of layer would control transform. In the case of statistical analysis in tamoxifen and in doxorubicin, the values of AUC were 0.58 and 0.64, respectively.
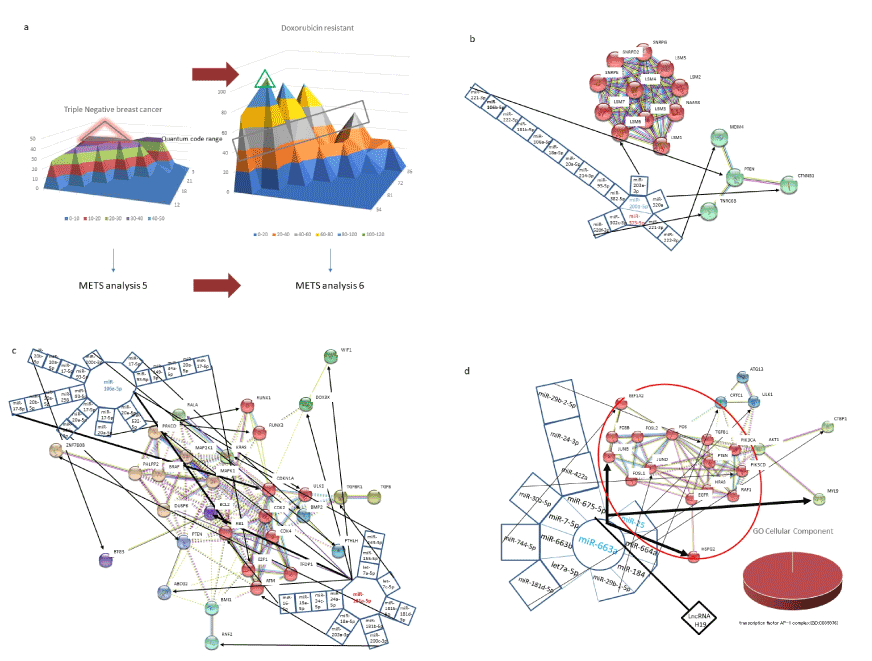
Figure 8.METS simulation of doxorubicin resistance in human breast cancer. Triple negative human breast cancer (METS analysis 5) was medicated with doxorubicin (a) and drug resistance was emerged (METS analysis 6). The QCR 40-50 was compared with each other. METS analysis 5. The triple negative breast cancer upregulated miR-373-5p and downregulated miR-200a-5p in QCR 40-50 (b). METS analysis 6.1. The network of doxorubicin resistant breast cancer was simulated to be compared with triple negative breast cancer in QCR 40-50 (c). The doxorubicin resistant breast cancer upregulated miR-181a-5p and downregulated miR-106a-5p. METS analysis 6.2. Doxorubicin resistant breast cancer was also simulated in QCR 90-100 (d). Red circled protein gene cluster (enclosed by a large red circle) was analyzed by GO enrichment and it showed as AP-1 complex.
Other chemotherapeutic agents against cancer
As described above, drug resistance was observed in not only immunotherapy but also chemotherapy against cancers. Cisplatin or carboplatin has been applied for therapy of cancers, such as non-small cell lung cancer, which is approximately 85% of lung cancer, however; drug resistance and decreasing drug sensitivity of chemotherapy exhibit and lead to poor efficacy of it. Fadejeva et al. have reported the summary of implication of miRNA in cisplatin-resistance of non-small cell lung cancer [108]. Cisplatin-resistance was associated with complex mechanisms, apoptosis, drug transport, proliferation, metastasis, DNA damage etc. Thirty-two miRNAs validated were assigned to apoptosis, and 22 to cell proliferation and cell cycle, 7 to DNA damage and 9 to drug transport were reviewed. In the case of oxaliplatin, miR-503-5p was upregulated in the drug resistant colorectal carcinoma [109]. Knockdown of miR-503-5p increased sensitivity to oxaliplatin, therefore miR-503-5p induced oxaliplatin resistance through inhibition of apoptosis by targeting the p53 upregulated modulator protein of apoptosis (PUMA). On the contrary, miR-128 overexpression suppressed paclitaxel-resistant non-small lung cancer stem cells and miR-107 enhanced paclitaxel-sensitivity of non-small lung cancer [110,111]. LncRNA MALAT1 knockdown reversed temozolomide resistance of glioma cells via miR-101 increasing [112]. Together, these data indicated that fail or success of chemotherapy to cancer would depend on alteration of miRNA expression in drug treatment. Thus, METS analysis would be useful to elucidate drug resistance.
Nutrients
Nutrients of diet could also affect miRNA expression. Quintanilha et al. have summarized the evaluated alteration of miRNAs by nutrients, which are not only polyphenol but also saturated fatty acid (SFA), PUFA, minerals, vitamins, and food derived miRNAs [113]. Shortly, SFA, palmitic acid increased miR-29a in human myocytes and miR-27b in mouse [114,115]. Eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) as PUFAs downregulated miR-146a, miR-146b, miR-21, miR-125a, and miR-155 [116]. Food sources of PUFA intervention also altered miRNA profile in the plasma [117]. Omega-3 PUFA, DHA downregulated miR-26a/b in human cholangiocarcinoma cells [118]. MiR-26a/b reduction resulted to inhibit the carcinoma cell proliferation via inhibiting the NAD+ dependent 15-hydroxyprostaglandin dehydrogenase (15-PGDH) through PGE2 inactivation, therefore, DHA prevented cholangiocarcinoma growth. In the case of rat model, ω-3 PUFA significantly downregulated miR-19b-3p, miR-146b-5p and miR-183-5p and suppressed inflammation [119]. Zanoaga et al. have summarized breast cancer connection between ω-3 or ω-6 PUFA and miRNA profiles. PUFA diets can affect miRNA expression, therefore, the environmental factors, such as nutrients have diverse effects against human health as a source of energy as well as genetic information modifier [120]. Vitamin D suppresses miR-155 expression that controls inflammation in patients [121]. Vitamin A (retinoic acid) upregulated miR-10a in breast cancer cells and upregulation of miR-10a suppresses breast cancer cells [122]. Vitamin C modulated miRNA expression of miR-3619-5p, miR-548a-3p, miR-4741, and miR-1825, miR-1208 in human bone marrow stromal cells [123]. Fatty acids and vitamin are nutrients, and some nutrients would bind to RNAs, however, fatty acids are oxidized in mitochondria to release energy and ascorbic acid (vitamin C) is a reducing agent during hydroxylation of collagen. It shows that two nutrients have distinct functions and miRNAs are also multiple functions, which would have the personal diversion, therefore, the relation among nutrients and miRNAs would be the multiple interactions based individual condition on demand. Quite recently, Gong et al. has found the difference expression profiles between American women of African ancestry and of European one, however, they could not determine whether triple negative breast cancer in African ancestry is more aggressive than European one [124]. It means that racing diversion would be linked to miRNA response, in turn, responses of miRNAs to therapeutic agent would also be involved in racing and personal diversion.
Chemotherapeutic and antibody anti-cancer agents, natural substances, nutrients and miRNA itself in foods affect expression of miRNAs in human. The responses of miRNA to chemicals and antibodies are distinct from in vitro pharmacokinetic reaction; therefore, chemical and antibody substances may have divergent characters in pharmacokinetics with side effect and drug resistance. In general, pharmaceutical company thinks that drugs would be designed not to be affected by personalized variations in pharmacokinetics and the main purpose for their tests would be establish common drug for good sale; however, alteration of miRNA profile means that the environmental substances have an important role for miRNA response to tune our lives and cancer therapy. Since the genetic test with miRNA can be achieved upon cheap and convenient form, a whole series of pharmaceutical drugs including ready commercial ones should be re-validated for personalized medicine by miRNA plus ncRNA expression profiles. Recently, clinical and experimental data have been supplied by open access journals for researchers to investigate miRNA biology, and the big databases of cancer, such as The Cancer Genome Atlas (TCGA) have also been launched into the net-covering the globe. Using these databases, the miRNA target prediction tool has been available for every researcher with equal conditions. We have introduced the quantum miRNA/miRNA interaction and the quantum language of miRNA was participated in the miRNA/target interaction on human cancers. Furthermore, the network of cancer among miRNA/miRNA/target genes, partly lncRNA has clearly been simulated by METS algorithm. On the contrary, drug resistance on approximate 1.4 million women of breast cancer patients is the serious problem in treatment of cancer, but the mechanisms of resistance are acquired, multiple, complex, and not mutually exclusive, and remained to be cleared. To understand the diversity of miRNA response against the chemo-substance, drug resistance in human breast cancer was shown to contextually simulated by METS with quantum multilayer diagram. Trastuzumab resistance simulation resulted significantly high value of AUC. However, neither of tamoxifen and doxorubicin miRNA/miRNA/target context was statistically enough to simulate the drug resistant state of breast cancer because lncRNA and circRNA data were absolute less than microRNA data. Although dynamic computer simulation models of drug resistance in breast cancer showed that in vivo pharmacokinetic effects of drug are much distinct from those of it in vitro, it is simultaneously suggested that miRNA gene expression would be altered by environmental quantum energy or its substances, such as chemicals, antibodies and nutrients. Thus, the METS simulation with quantum language of miRNA would contribute to develop prediction of resistant state, recurrence of cancer and new cancer precious medicine. The quantum language of miRNA against foods and their contents, and drug administration according divergent might be useful for human lives to increase the mortality rate of cancers by inhibition of the incidence on drug resistance, finally by anti-cancer.
The author declare not having conflicts of interest.
View Supplementary Data
- Wilusz JE, Sunwoo H, Spector DL (2009) Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 23: 1494-1504. [crossref]
- Yoshikawa M, Fujii YR (2016) Human ribosomal RNA-derived resident microRNAs as the transmitter of information upon the cytoplasmic cancer stress. Biomed Res Int 2016: 1-14.
- Green D Fraser WD, Dalmay T (2016) Transfer RNA-derived small RNAs in the cancer transcriptome. Pflugers Arch 468: 1041-1047. [crossref]
- Wang KC, Chang HY (2011) Molecular mechanisms of long noncoding RNAs. Mol Cell 43: 904-914. [crossref]
- Bezzi M, Guarnerio J, Pandolfi PP (2017) A circular twist on microRNA regulation. Cell Res 27: 1401-1402. [crossref]
- Holdt LM, Kohlmaier A, Teupser D (2018) Molecular roles and function of circular RNAs in eukaryotic cells. Cell Mol Life Sci 75: 1071-1098. [crossref]
- Fujii YR (2017) The microRNA 2000: from HIV-1 to healthcare. Scientific Research Publishing, Inc., Irvine, CA.
- Klinge CM (2018) Non-coding RNAs: long non-coding RNAs and microRNAs in endocrine-related cancers. Endocr Relat Cancer 25: R259-259R282. [crossref]
- Campos-Parra AD, Mitznahuatl GC, Pedroza-Torres A, Romo RV, Reyes FIP, et al. (2017) Micro-RNAs as potential predictors of response to breast cancer systemic therapy: future clinical implication. Int J Mol Sci 18: 1182. [crossref]
- Wu XS, Wang F, Li HF, et al. (2017) LncRNA-PAGBC acts as a microRNA sponge and promotes gallbladder tumorigenesis. EMBO Rep 18: 1837-1853. [crossref]
- Gong LC, Xu HM, Guo GL, Zhang T, Shi JW, et al. (2017) Long non-coding RNA H19 protects H9c2 cells against hypoxia-inducing injury by targeting microRNA-139. Cell Physiol Biochem 44: 857-869. [crossref]
- Wu Q, Meng WY, Jie Y, Zhao H (2018) LncRNA MALAT1 induces colon cancer development by regulating miR-129-5p/HMGB1 axis. J Cell Physiol 233: 6750-6757. [crossref]
- Wang KC, Chang HY (2017) Transcription coactivator and lncRNA duet evoke Hox genes. PLoS Genet 13: e1006797. [crossref]
- Liz J, Portela A, Soler M, Gómez A, Ling H, et al. (2014) Regulation of pri-miRNA processing by a long noncoding RNA transcribed from an ultraconserved region. Mol Cell 55: 138-147. [crossref]
- Han C, Seebacher NA, Hornicek FJ, Kan Q, et al. (2017) Regulation of microRNAs function by circular RNAs in human cancer. Oncotarget 8: 64622-64637. [crossref]
- Li G, Yang H, Han K, Zhu D, Lun P, et al. (2018) A novel circular RNA, hsa_circ_0046701, promotes carcinogenesis by increasing the expression of miR-142-3p target ITGB8 in glioma. Biochem Biophys Res Commun 498: 254-261. [crossref]
- Cheng Z, Li Z, Ma K, Li X, Tian N, et al. (2017) Long Non-coding RNA XIST Promotes Glioma Tumorigenicity and Angiogenesis by Acting as a Molecular Sponge of miR-429. J Cancer 8: 4106-4116. [crossref]
- Yu H, Xue Y, Wang P, Liu X, Ma J, et al. (2017) Knockdown of long non-coding RNA XIST increases blood-tumor barrier permeability and inhibits glioma angiogenesis by targeting miR-137. Oncotargets 6: e303. [crossref]
- Ma Y, Wang P, Xue Y, Qu C, Zheng J, et al. (2017) PVT1 affects growth of glioma microvascular endothelial cells by negatively regulating miR-186. Tumor Biol 2017: 1-8. [crossref]
- Gao Y, Yu H, Liu Y, Liu X, Zheng J, et al. (2018) Long non-coding RNA HOXA-AS2 regulates malignant glioma behaviors and vasculogenic mimicry formation via the miR-373/EGFR axis. Cell Phys Biochem 45: 131-147. [crossref]
- Wang G, Li Z, Tian N, Han L, Fu Y, et al. (2016) miR-148b-3p inhibits malignant biological behaviors of human glioma cells induced by high HOTAIR expression. Onc Let 12: 879-886. [crossref]
- Bian EB, Ma CC, He XJ, et al. (2016) Epigenetic modification of miR-141 regulates SKA2 by an endogenous 'sponge' HOTAIR in glioma. Oncotarget 7: 30610-30625. [crossref]
- Shi C, Yang Y, Yu J, Meng F, Zhang T, et al. (2017) The long noncoding RNA LINC00473, a target of microRNA 34a, promotes tumorigenesis by inhibiting ILF2 degradation in cervical cancer. Am J Cancer Res 7: 2157-2168. [crossref]
- Wang L, Bu P, Ai Y, Srinivasan T, Chen HJ, et al. (2016) A long non-coding RNA targets microRNA miR-34a to regulate colon cancer stem cell asymmetric division. eLife 5: e14620. [crossref]
- Cheng C, Qin Y, Zhi Q, Wang J, Qin C (2018) Knockdown of long non-coding RNA HOTAIR inhibits cisplatin resistance of gastric cancer cells through inhibiting the PI3K/Akt and Wnt/ß-catenin signaling pathways by up-regulating miR-34a. Int J Biol Macromol 107: 2620-2629. [crossref]
- Chiyomaru T, Yamamura S, Fukuhara S, Yoshino H, Kinoshita T, et al. (2013) Genistein inhibits prostate cancer cell growth by targeting miR-34a and oncogenic HOTAIR. PLoS One 8: e70372. [crossref]
- Campos-Parra AD, López-Urrutia E, Orozco Moreno LT, López-Camarillo C, Meza-Menchaca T, et al. (2018) Long non-coding RNAs as new master regulators of resistance to systemic treatments in breast cancer. Int J Mol Sci 19: 2711. [crossref]
- Yang Y, Fan X, et al. (2017) Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res 27: 626-641. [crossref]
- Mohr AM, Mott JL (2015) Overview of microRNA biology. Semin Liver Dis 35: 3-11. [crossref]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP (2011) A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146: 353-358. [crossref]
- Fujii YR (2013) The RNA gene information: retroelement-microRNA entangling as the RNA quantum code. Methods Mol Biol 936: 47-67.
- Bo L, Wei B, Wang Z, Kong D, Gao Z, et al. (2017) Identification of key genes in glioma CpG island methylator phenotype via network analysis of gene expression data. Mol Med Rep 16: 9503-9511. [crossref]
- Cao Y, Wang P, Ning S, Xiao W, Li X (2016) Identification of prognostic biomarkers in glioblastoma using a long non-coding RNA-mediated, competitive endogenous RNA network. Oncotarget 7: 41737-41747. [crossref]
- Chiu HS, Llobet-Navas D, Yang X, Chung WJ, Ambesi-Impiombato A, et al. (2018) Cupid: simultaneous reconstruction of microRNA-target and ceRNA network. Gen Res 25: 257-267. [crossref]
- Li J, Zhang Q, Fan X, Mo W, Dai W, et al. (2017) The long noncoding RNA TUG1 acts as a competing endogenous RNA to regulate the Hedgehog pathway by targeting miR-132 in hepatocellular carcinoma. Oncotarget 8: 65932-65945. [crossref]
- Piwecka M, Glažar P, Hernandez-Miranda LR, Memczak S, Wolf SA, et al. (2017) Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357: 6357. [crossref]
- Hentze MW, Preiss T (2013) Circular RNAs: splicing's enigma variations. EMBO J 32: 923-925. [crossref]
- Xia B, Wang L, Feng L, Tian B, Tan Y, et al. (2018) Knockdown of long non-coding RNA CAT104 inhibits the proliferation, migration and invasion of human osteosarcoma cells by regulating microRNA-381. Onc Res
- Fujii YR, Saksena NK (2008) Viral infection-related microRNAs in viral and host genomic evolution. In: Morris KV, (eds.) RNA and the regulation of gene expression. Horizon Scientific Press, London.
- Yoshikawa M, Osone T, Fujii YR (2016) MicroRNA memory I: the positive correlation between synergistic effects of microRNAs in cancer and a novel quantum scoring system. J Adv Med Phar Sci 5: 1-16.
- Zhao Q, Liu Y, Zhang N, Hu M, Zhang H, et al. (2018) Evidence for plant-derived xenomiRs based on a large-scale analysis of public small RNA sequencing data from human samples. PLoS One 13: e0187519. [crossref]
- Li Z, Xu R, Li N (2018) MicroRNAs from plants to animals, do they define a new messenger for communication? Nut Meta 15: 68. [crossref]
- Manca S, Upadhyaya B, Mutai E, Desaulniers AT, Cederberg RA, et al. (2018) Milk exosomes are bioavailable and distinct microRNA cargos have unique tissue distribution patterns. Sci Rep 8: 11321.
- Lukasik A, Brzozowska I, Zielenkiewicz U, Zielenkiewicz P4 (2017) Detection of Plant miRNAs Abundance in Human Breast Milk. Int J Mol Sci 19. [crossref]
- Fujii YR (2018) Quantum language of microRNA: application for new cancer therapeutic targets. Methods Mol Biol 1733: 145-157. [crossref]
- Osone T, Yoshikawa M, Fujii YR (2016) MicroRNA memory II: a novel scoring integration model for prediction of human disease by microRNA/microRNA quantum multi-interaction. J Adv Med Phar Sci 5: 1-18.
- Hill M, Tran N (2018) MicroRNAs Regulating MicroRNAs in Cancer. Trends Cancer 4: 465-468. [crossref]
- Ning P, Wu Z, Hu A, Li X, He J, et al. (2018) Integrated genomic analyses of lung squamous cell carcinoma for identification of a possible competitive endogenous RNA network of TCGA datasets. Peer J 6: e4254. [crossref]
- Yao K, Wang Q, Jia J, Zhao H (2017) A competing endogenous RNA network identifies novel mRNA, miRNA and lncRNA markers for the prognosis of diabetic pancreatic cancer. Tumor Biol 2017: 1-13. [crossref]
- Xue WH, Fan ZR, Li LF, Lu JL, Ma BJ, et al. (2018) Construction of an oesophageal cancer-specific ceRNA network based on miRNA, lncRNA, and mRNA expression data. World J Gastro 24: 23-34. [crossref]
- Xu JH, Chang WH, Fu HW, Yuan T, Chen P (2018) The mRNA, miRNA and lncRNA networks in hepatocellular carcinoma: an integrative transcriptomic analysis from gene expression omnibus. Mol Med Rep. 17: 6472–6482. [crossref]
- Li F, Li Q, Wu X (2018) Construction and analysis for differentially expressed long non-coding RNAs and microRNAs mediated competing endogenous RNA network in colon cancer. PLoS One 13: e0192494. [crossref]
- Zhang Z, Wang S, Ji D, Qian W, Wang Q, et al. (2018) Construction of a ceRNA network reveals potential incRNA biomarkers in rectal adenocarcinoma. Oncol Rep 39: 2101-2113. [crossref]
- Song Z, Zhao W, Cao D, Zhang J, Chen S (2018) Elementary screening of lymph node metastatic-related genes in gastric cancer based on the co-expression network of messenger RNA, microRNA and long non-coding RNA. Br J Med Biol Res 51: e6685. [crossref]
- Lu M, Xu X, Xi B, Dai Q, Li C, et al. (2018) Molecular network-based identification of competing endogenous RNAs in thyroid carcinoma. Genes 9: 44. [crossref]
- Li MP, Hu YD, Hu XL, et al. (2016) MiRNAs and miRNA Polymorphisms Modify Drug Response. Int J Environ Res Public Health 13. [crossref]
- Liu T, Wu C, Weng G, Zhao Z, He X, et al. (2017) Bufalin inhibits cellular proliferation and cancer stem cell-like phenotypes via upregulation of miR-203 in glioma. Cell Physiol Biochem 44: 671-681. [crossref]
- Chang Y, Zhao Y, Gu W, Cao Y, Wang S, et al. (2015) Bufalin inhibits the differentiation and proliferation of cancer stem cells derived primary osteosarcoma cells through miR-148a. Cell Physiol Biochem 36: 1186-1196. [crossref]
- Guo X, Yu L, Chen M, Wu T, Peng X, et al. (2016) miR-145 mediated the role of aspirin in resisting VSMCs proliferation and anti-inflammation through CD40. J Transl Med 14: 211. [crossref]
- LeMay-Nedjelski L, Mason-Ennis JK, Taibi A, Comelli EM, Thompson LU (2018) Omega-3 polyunsaturated fatty acids time-dependently reduced cell viability and oncogenic microRNA-21 expression in estrogen receptor-positive breast cancer. Int J Mol Sci 19: 244. [crossref]
- Shi H, Bi H, Sun X, Dong H, Jiang Y, et al. (2018) Antitumor effects of Tubeimoside-1 in NCI-H1299 cells are mediated by microRNA-126-5p-induced inactivation of VEGF-A/VEGFR-2/ERK signaling pathway. Mol Med Rep 17: 4327-4336. [crossref]
- Du M, Zhang Z, Gao T (2017) Piceatannol induced apoptosis through up-regulation of micriRNA-181a in melanoma cells. Biol Res 50: 36.
- Tili E, Michaille JJ (2016) Promiscuous effects of some phenolic natural products on inflammation at least in part arise from their ability to modulate the expression of global regulators, namely microRNAs. Molecules 21: 1263.
- Ahmad A, Li Y, Bao B, Kong D, Sarkar FH (2014) Epigenetic regulation of miRNA-cancer stem cells nexus by nutraceuticals. Mol Nutr Food Res 58: 79-86. [crossref]
- Lamichhane TN, Leung CA, Douti LY, Jay SM (2017) Ethanol induces enhanced vascularization bioactivity of endothelial cell-derived extracellular vesicles via regulation of microRNAs and long noncoding RNAs. Sci Rep 7: 13794.
- Nosrati N, Bakovic M, Paliyath G (2017) Molecular mechanisms and pathways as targets for cancer prevention and progression with dietary compounds. Int J Mol Sci 18. [crossref]
- Alehagen U, Johansson P, Aaseth J, Alexander J, Wågsäter D (2017) Significant changes in circulating microRNA by dietary supplementation of selenium and coenzyme Q10 in healthy elderly males. A subgroup analysis of a prospective randomized double-blind placebo-controlled trial among elderly Swedish citizens. PLoS One 12: e0174880.
- O'Day E, Lal A (2010) MicroRNAs and their target gene networks in breast cancer. Breast Cancer Res 12: 201. [crossref]
- Martin HL, Smith L, Tomlinson DC (2014) Multidrug-resistant breast cancer: current perspectives. Breast Cancer 6: 1-13.
- Mandujano-Tinoco EA, García-Venzor A, Melendez-Zajgla J, Maldonado V (2018) New emerging roles of microRNAs in breast cancer. Breast Cancer Res Treat 171: 247-259. [crossref]
- Adams BD, Furneaux H, White BA (2007) The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-a (ERa) and represses ERa messenger RNA and protein expression in breast cancer cell lines. Mol Endcrin 21: 1132-1147.
- Leivonen SK, Sahberg KK, Mäkelä R, Due EU, Kallioniemi O, et al. (2013) High-throughput screen identify microRNAs essential for HER2 positive breast cancer cell growth. Mol Onc 8: 93-104.
- Ye X, Bai W, Zhu H, Zhang X, Chen Y, et al. (2014) MiR-221 promotes trastuzumab-resistance and metastasis in HER2-positive breast cancers by targeting PTEN. BMB Rep 47: 268-273.
- Di Leva G, Cheung DG, Croce CM (2015) miRNA clusters as therapeutic targets for hormone-resistant breast cancer. Expert Rev Endocrinol Metab 10: 607-617.
- Rizzo S, Cangemi A, Galvano A, Fanale D, Buscemi S, et al. (2017) Analysis of miRNA expression profile induced by short term starvation in breast cancer cells treated with doxorubicin. Oncotarget 8: 71924-71932.
- Kahraman M, Röske A, Laufer T, Fehlmann T, et al. (2018) MicroRNA in diagnosis and therapy monitoring of early-stage triple-negative breast cancer. Sci Rep 8: 11584. [crossref]
- Zwart W, Griekspoor A, Berno V, Lakeman K, Jalink K, et al. (2007) PKA-induced resistance to tamoxifen is associated with an altered orientation of ERalpha towards co-activator SRC-1. EMBO J 26: 3534-3544. [crossref]
- Sun M, Paciga JE, Feldman RI, Yuan ZQ, Coppola D, et al. (2001) Phosphatidylinositol-3-OH kinase (PI3K)/AKT2, activated breast cancer, regulates and is induced by estrogen receptor a (ERa) via interaction between ERa and PI3K. Cancer Res 61: 5985-5991.
- Tomlinson DC, Knowles MA, Speirs V (2012) Mechanisms of FGFR3 actions in endocrine resistant breast cancer. Int J Cancer 130: 2857-2866. [crossref]
- Fox EM, Miller TW, Balko JM, Kuba MG, Sánchez V, et al. (2011) A genome-wide screen identifies the insulin/IGF-1 receptor pathway as a mechanism of escape from hormone dependence in breast cancer. Cancer Res 71: 6773-6784.
- Jung EJ, Santarpia L, Kim J, Esteva FJ, Moretti E, et al. (2012) Plasma microRNA 210 levels correlate with sensitivity to trastuzumab and tumor presence in breast cancer patients. Cancer 118: 2603-2614.
- Le XF, Almeida MI, Mao W, Spizzo R, Rossi S, et al. (2012) Modulation of MicroRNA-194 and cell migration by HER2-targeting trastuzumab in breast cancer. PLoS One 7: e41170. [crossref]
- Bai WD, Ye XM, Zhang MY, Zhu HY, Xi WJ, et al. (2014) MiR-200c suppresses TGF-ß signaling and counteracts trastuzumab resistance and metastasis by targeting ZNF217 and ZEB1 in breast cancer. Int J Cancer 135: 1356-1368.
- Lu X, Ma J, Chu J, Shao Q, Zhang Y, et al. (2017) MiR-129-5p Sensitizes the Response of Her-2 Positive Breast Cancer to Trastuzumab by Reducing Rps6. Cell Physiol Biochem 44: 2346-2356. [crossref]
- Sui M, Jiao A, Zhai H, Wang Y, Wang Y, et al. (2017) Upregulation of miR-125b is associated with poor prognosis and trastuzumab resistance in HER2-positive gastric cancer. Exp Ther Med 14: 657-663. [crossref]
- Shi SJ, Wang LJ, Yu B, Li YH, Jin Y, et al. (2015) LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget 6: 11652-11663.
- Li W, Zhai L, Wang H, Liu C, Zhang J, et al. (2016) Downregulation of lncRNA GAS5 causes trastuzumab resistance in breast cancer. Oncotarget 7: 27778-27786.
- De Mattos-Arruda L, Bottai G, Nuciforo PG, Di Tommaso L, Giovanetti E, et al. (2015) MicroRNA-21 links epithelial-to-mesenchymal transition and inflammation signals to confer resistance to neoadjuvant trastuzumab and chemotherapy in HER2-positive breast cancer patients. Oncotarget 6: 37269-37280.
- Trontti K, Väänänen J, Sipilä T, Greco D, Hovatta I (2018) Strong conservation of inbred mouse strain miRNA loci but broad variation in brain microRNAs due to RNA editing and isomiR expression. RNA 24: 643-655.
- Cilek EE, Ozturk H, Dedeoglu BG (2017) Construction of miRNA-miRNA networks revealing the complexity of miRNA-mediated mechanisms in trastuzumab treated breast cancer cell lines. PLoS One 12: e0185558.
- Hu W, Tan C, et al. (2018) Functional miRNAs in breast cancer drug resistance. Onco Targets Ther 11: 1529-1541. [crossref]
- Yu Z, Wang C, Wang M, Li Z, Casimiro MC, et al. (2008) A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J Cell Biol 182: 509-517. [crossref]
- Li Y, Hong F, Yu Z (2012) Decreased expression of microRNA-206 in breast cancer and its association with disease characteristics and patient survival. J Int Med Res 41: 596-602.
- Samaeekia R1, Adorno-Cruz V2,3, Bockhorn J1,4, et al. (2017) miR-206 Inhibits Stemness and Metastasis of Breast Cancer by Targeting MKL1/IL11 Pathway. Clin Cancer Res 23: 1091-1103. [crossref]
- Zhao G, Guo J, Li D, Jia C, Yin W, et al. (2013) MicroRNA-34a suppresses cell proliferation by targeting LMTK3 in human breast cancer mcf-7 cell line. DNA Cell Biol 32: 699-707. [crossref]
- Eichelser C, Flesch-Janys D, Chang-Claude J, Pantel K, Schwarzenbach H (2013) Deregulation serum concentrations of circulating cell-free microRNAs miR-17, miR-34a, miR-155, and miR-373 in human breast cancer development and progression. Clin Chem 59: 1489-1496.
- Mansoori B, Mohammadi A, Shirjang S, Baghbani E, Baradaran B (2016) Micro RNA 34a and let-7a expression in human breast cancers is associated with apoptotic expression genes. Asian Pac J Cancer Prev 17: 1887-1890.
- Hynes NE, MacDonald G (2009) ErbB receptors and signaling pathways in cancer. Curr Opin Cell Biol 21: 177-184. [crossref]
- Kulkarni S, Dolezal JM, Wang H, Jackson L, Lu J, et al. (2017) Ribosomopathy-like properties of murine and human cancers. PLoS One 12: e0182705. [crossref]
- Marty M, Cognetti F, Maraninchi D, Snyder R, Mauriac L, et al. (2005) Randomized phase II trial of the efficacy and safety of trastuzumab combined with docetaxel in patients with human epidermal growth factor receptor 2-positive metastatic breast cancer administered as a first-line treatment: the M77001 study group. J Clin Oncol 23: 4265-4274.
- Zou Q, Yi W, Huang J1 Fu F, Chen G, et al. (2017) MicroRNA-375 targets PAX6 and inhibits the viability, migration and invasion of human breast cancer MCF-7 cells. Exp Ther Med 14: 1198-1204. [crossref]
- Cui YX, Bradbury R, Flamini V, Wu B, Jordan N, et al. (2017) MicroRNA-7 suppresses the homing and migration potential of human endothelial cells to highly metastatic human breast cancer. Bri J Cancer 117: 89-101.
- Block I, Burton M, Sorensen KP, Andersen L, Larsen MJ, et al. (2018) Association miR-548c-5p, miR-7-5p, miR-210-3p, miR-128-3p with recurrence in systematically untreated breast cancer. Oncotarget 9: 9030-9042.
- Ye XM, Zhu HY, Bai WY, Wang T, Wang L, et al. (2014) Epigenetic silencing of miR-375 induces trastuzumab resistance in HER2-positive breast cancer by targeting IGF1R. BMC Cancer 14: 134.
- Huynh FC, Jones FE (2014) MicroRNA-7 inhibits multiple oncogenic pathways to suppress HER2?16 mediated breast tumorigenesis and reverse trastuzumab resistance. PLoS One 9: e114419.
- Yang ZQ, Liu G, Bollig-Fischer A, Giroux CN, Ethier SP (2010) Transforming properties of 8p11-12 amplified genes in human breast cancer. Mol Cell Pathol 70: 8487-8479.
- Wang J, Wang X, Chen T, Jiang L, Yang Q (2017) Huaier extract inhibits breast cancer progression through a lncRNA-H19/miR-675-5p pathway. Cell Physiol Biochem 44: 581-593.
- Fadejeva I, Olschewski H, Hrzenjak A1 (2017) MicroRNAs as regulators of cisplatin-resistance in non-small cell lung carcinomas. Oncotarget 8: 115754-115773. [crossref]
- Xu K, Chen G, Qiu Y, et al. (2017) miR-503-5p confers drug resistance by targeting PUMA in colorectal carcinoma. Oncotarget 8: 21719-21732. [crossref]
- Koh H, Park H, Chandimali N, Huynh DL, Zhang JJ, et al. (2017) MicroRNA-128 suppresses paclitaxel-resistant lung cancer by inhibiting MUC1-C and BMI-1 in cancer stem cells. Oncotarget 8: 110540-110551. [crossref]
- Lu C, Xie Z, Peng Q (2017) MiRNA-107 enhances chemosensitivity to paclitaxel by targeting antiapoptotic factor Bcl-w in non-small cell lung cancer. Am J Cancer Res 7: 1863-1873.
- Cai T, Liu Y, Xiao J (2018) Long noncoding RNA MALAT1 knockdown reverses chemoresistance to temozolomide via promoting microRNA-101 in glioblastoma. Cancer Med 7: 1404-1415. [crossref]
- Quintanilha BJ, Reis BZ, Duarte S, Cozzolino SM, Rogero MM (2017) Nutrimiromics: Role of microRNAs and nutrition in modulating inflammation and chronic diseases. Nutrients 2017: 9.
- Yang WM, Jeong HJ, Park SY, Lee W (2014) Induction of miR-29a by saturated fatty acids impairs insulin signaling and glucose uptake through translational repression of IRS-1 in myocytes. FEBS Let 588: 2170-2176.
- Takahashi K, Sasano T, Sugiyama K, Kurokawa J, Tamura N, et al. (2016) High-fat diet increases vulnerability to atrial arrhythmia by conduction disturbance via miR-27b. J Mol Cell Cardiol 90: 38-46. [crossref]
- Roessler C, Kuhlmann K, Hellwing C, Leimert A, Schumann J (2017) Impact of polyunsaturated fatty acids on miRNA profiles of monocytes/macrophages and endothelial cells. Int J Mol Sci 18: 284.
- Ortega FJ, Cardona-Alvarado MI, Mercader JM, Moreno M, Sabater M, et al. (2015) Circulating profiling reveals the effect of a polyunsaturated fatty acid-enriched diet on common miroRNAs. J Nutr Biochem 26: 1095-1101.
- Yao L, Han C, Song K, Zhang J, Lim K et al. (2015) ?-3 polyunsaturated fatty acids up-regulate the expression of 15-PGDH by inhibiting miR-26a and miR-26b in human cholangiocarcinoma cells. Cancer Res 75: 1388-1398.
- Zheng Z, Ge Y, Zhang J, Xue M, Li Q, et al. (2015) PUFA diets alter the microRNA expression profiles in an inflammation rat model. Mol Med Rep 11: 4149-4157. [crossref]
- Zanoaga O, Jurj A, Raduly L, Cojocneanu-Petric R, et al. (2018) Implications of dietary ω-3 and ω-6 polyunsaturated fatty acids in breast cancer. Exp Ther Med 15: 1167-1176. [crossref]
- Kempinska-Podhorodecka A, Milkiewicz M, Wasik U, Ligocka J, Zawadzki M, et al. (2017) Decreased expression of vitamin D receptor affects an immune response in primary biliary cholangitis via the VDR-miR155-SOCS1 pathway. Int J Mol Sci 18: 289.
- Khan S, Wall D, Curran C, Newell J, Kerin MJ, et al. (2015) MicroRNA-10a is reduced in breast cancer and regulated in part through retinoic acid. BMC Cancer 15: 345. [crossref]
- Kolhe R, Mondal AK, Pundkar C, Periyasamy-Thandavan S, Mendhe B, et al. (2018) Modulation of miRNAs by Vitamin C in Human Bone Marrow Stromal Cells. Nutrients 10. [crossref]
- Gong Z, Wang J, Wang D, Buas MF, Ren X, et al. (2018) Differences in microRNA expression in breast cancer between women of African and European ancestry. Carcinogenesis 134.
- Ma T, Lu Y, Zhang J (2015) miRNA-542-3p downregulation promotes trastuzumab resistance in breast cancer cells via AKT activation. Onc Rep 33: 1215-1220.
- Eto K, Iwatsuki M, Watanabe M, Ida S, Ishimoto T, et al. (2014) The microRNA-21/PTEN pathway regulates the sensitivity of HER2-positive gastric cancer cells to trastuzumab. Ann Surg Oncol 21: 343-350. [crossref]
- De Cola A, Volpe S, Budani MC, Ferracin M, Lattanzio R, et al. (2015) miR-205-5p-mediated downregulation of ErbB/HER receptors in breast cancer stem cells results in targeted therapy resistance. Cell Death Dis 6: e1823.
- Pardo O, Castellano L, Munro CE, Hu Y, Mauri F, et al. (2016) miR-515-5p controls cancer cell migration through MARK4 regulation. EMBO Rep 17: 570-584. [crossref]