Background: Early detection of colorectal adenoma plays crucial roles in CRC early prevention and intervention. Current detection by colonoscopy is not suitable for mass screening due to its high costs and low compliance. The SEPT9 gene methylation assay is a new qPCR-based assay for CRC screening, and was found to be effective for adenoma detection. However, its ability in detecting adenoma has not been fully examined.
Methods: We performed a case-control study and investigated the relationship between the plasma level of methylated SEPT9 (mSEPT9) and the sensitivity of colorectal adenoma detection. The plasma mSEPT9 level was measured from subjects with confirmed diagnosis of various types of adenomas. Plasma samples from normal subjects and patients with polyps or CRC were also measured for comparison.
Results: It was found that the detection sensitivity was positively correlated with pathological severity and the level of plasma mSEPT9 in linear relationship. Adenomas with villous components or high-grade dysplasia (HGD) exhibited higher mSEPT9 level and higher sensitivity than those without villous components or HGD.
Conclusion: Our study suggests that the detection sensitivity of the mSEPT9 assay in colorectal adenoma was determined by the plasma level of mSEPT9, which reflects the severity of lesions.
Septin 9, SEPT9, methylation, colorectal cancer, adenoma, dysplasia
Colorectal cancer (CRC) is the third most common cancer in the world [1]. Screening of CRC can reduce mortality and prolong the survival time. Current CRC screening methods, such as fecal immunochemical test (FIT), FIT-DNA test or the mSEPT9 DNA test, were designed to detect early-stage CRC and can detect some precancerous lesions as well. Detection of CRC reduces mortality while detection of precancerous lesions can reduce incidence. Therefore, screening aiming at precancerous lesions has great potential in cancer prevention. However, detection of precancerous lesions in screening using current methods is accompanied by high false positive rate in other colorectal diseases and normal subjects, which hinders the use of screening methods, and further confirmatory diagnosis, such as colonoscopy, is still required.
The mSEPT9 test is an assay approved by the US FDA as a CRC screening test for average-risk population from 50 to 75 years old. It can detect 68% of CRC with a specificity of 80% in screening of average-risk population [2,3]. Generally speaking, it showed a sensitivity from 48.2% to 95.6% with a specificity from 79.1% to 99.1% in CRC detection [4]. Previous studies reported that the detection rate of the mSPET9 test for adenoma and hyperplastic polyps were much lower than that for CRC [2-9]. However, our recent study found that the mSEPT9 test is capable of detecting adenomas with villous components or high-grade dysplasia (HGD) with adequate sensitivity using the 1/3 algorithm [9].
In this study, we investigated the relationship between the sensitivity of adenoma detection and the plasma level of mSEPT9. We found that the sensitivity was positively correlated with the level of mSEPT9 in plasma in linear relationship, and the level of mSEPT9 in plasma was also correlated with the severity of colorectal diseases. It was also found that the mSEPT9 level in adenomas with villous components or HGD was higher than those without villous components or HGD, indicating that these two factors led to high level of mSEPT9 and may be high-risk factors in adenoma. Our observation strongly suggests that the plasma level of mSEPT9 is an indicator for disease severity and detection sensitivity, and the assay itself is effective in adenoma detection.
Ethics
The plan for the study was submitted to the ethics committee of the participating hospitals for review and approval before the start of the clinical study. All subjects signed an informed consent before blood collection, and they were informed of the usage of plasma and the test results. Confirmation of approval for clinical studies was received from all named institutional review boards or ethics committees.
The case control study was designed and implemented in three Chinese hospitals using the Epi proColon 2.0 CE assay (Epigenomics AG, Berlin, Germany). Clinical status was determined before blood draw for the SEPT9 assay, and blood samples were obtained from all subjects who met the selection criteria. All technicians were blinded to the clinical information of subjects. A total of 484 subjects was recruited in this study, including 68 CRC patients, 173 subjects with adenomas, 108 subjects with hyperplastic polyps (abbreviated as ‘polyps’ or HP in all tables and figures), and 135 subjects with no evidence of disease (NED) (Table 1). The classification of all conditions was based on diagnosis from colonoscopy and subsequent pathological examinations. All subjects were divided into four subgroups based on disease status. Patients with adenoma were divided into serrated (SA), tubular (TA), tubulovillous (TVA) and villous adenoma (VA) groups based on the results from pathological examinations. None of the subjects received chemotherapy, radiotherapy, or surgical intervention before the blood draw and the colonoscopy examination.
Table 1. The number of enrolled subjects by pathological diagnostic group
Diagnosis group |
Total |
Gender |
Age |
Male |
Female |
<50 |
50-59 |
60-69 |
≥ 70 |
CRC |
68 |
43 |
25 |
8 |
19 |
27 |
14 |
Adenoma |
173 |
111 |
62 |
37 |
59 |
46 |
31 |
serrated |
15 |
9 |
6 |
2 |
5 |
6 |
2 |
Tubular |
101 |
74 |
27 |
26 |
37 |
23 |
15 |
Tubulovillous |
54 |
23 |
31 |
7 |
16 |
19 |
12 |
Villous |
18 |
14 |
4 |
4 |
6 |
4 |
4 |
HP |
108 |
77 |
31 |
42 |
42 |
13 |
11 |
NED |
135 |
66 |
69 |
60 |
43 |
23 |
9 |
CRC=Colorectal Cancer; HP=hyperplastic polyps; NED=No Evidence of Diseases
Sample size estimation
Sample size estimation was based on the following equation for known positive detection rate: N=Z2*[p (1-p)]/E2. The parameters were defined as follows: Z is a statistical parameter (Z=1.96 for 95% CI); E represented the error (10% was chosen in this study), and p represented the putative positive detection rate). The p value (0.35) was obtained from a previous pilot study looking at the sensitivity of Epi proColon 2.0 CE assay on adenoma. From this, an estimated 88 adenoma cases were required. From the estimation that adenoma accounts for 30% of all patients, at least 293 patients should be included; therefore, the study goal was to recruit 367 patients, anticipating a 20% loss of follow-up rate (Table 1).
Sample collection and storage
Samples were collected from outpatients and inpatients, and the sample information was recorded in sample collection forms. A 10 ml peripheral blood sample was collected with 10 ml K2EDTA anticoagulant tubes (BD biosciences, Franklin Lakes, NJ, USA) to ensure the accuracy of the assay. Sample storage and transportation followed the instructions for use of the Epi proColon 2.0 CE assay.
Plasma samples from all three hospitals were prepared in individual hospitals and stored under -20 ℃ before they were delivered to Beijing BioChain Medical Laboratory, and all assays were performed in the same laboratory within three weeks from the sample collection date. The sample quality was examined when the samples arrived at the medical laboratory. Samples with plasma volume less than 3.5 ml, or with apparent hemolysis, high bilirubin, chylemia, or visible particles or pellets were not tested, and repeated blood draw was requested.
qPCR analysis of SEPT9
DNA extraction from plasma samples and bisulfite conversion were performed manually strictly following the manufacturer’s instructions of Epi proColon 2.0 CE assay. The bisDNA was assayed with Epi proColon 2.0 CE kits on an ABI7500 Fast Dx Real Time PCR device (Life Technologies).
Data analysis and interpretation
The data from the PCR reactions of the Epi proColon 2.0 CE assay were analyzed using the 1/3 algorithm, which means that a sample was considered to be positive if at least one of the three PCR replicates was positive and was considered to be negative if all three PCR replicates were negative. Statistical analysis was performed and the receiver operating characteristic (ROC) curves were plotted with Graphpad Prism 5.0 software (GraphPad Software, Inc, La Jolla, CA 92037, USA). For each sample, a relative methylation value was determined using the ΔΔCt method adapted for DNA methylation analyses as previously described [10]. In brief, ΔΔCt values were calculated as below:
ΔΔCtSample=ΔCtSample-ΔCTCalibrator, where ΔCtSample=CtACTB of Sample-CtSEPT9 of Sample and ΔCtCalibrator=CtACTB of Calibrator-CtSEPT9 of Calibrator.
Plasma mSEPT9 level reflected the severity of colorectal diseases and determined the detection sensitivity
We first investigated the sensitivity of the mSEPT9 assay in normal subjects, and patients with hyperplastic polyps (HP), adenoma and CRC. We also analyzed the plasma mSEPT9 levels of all subjects from the above groups. It shows from the box and whisker plot in Figure 1A that the levels of plasma mSEPT9 (ΔΔCt) in HP and adenoma exhibited no significant differences to that of the normal subjects. In contrast, significant increase in mSEPT9 level was observed in CRC groups compared with other groups. The sensitivity and the mean values of mSEPT9 levels from all the above groups were plotted in Figure 1B. It can be observed that the sensitivity increased with the elevation of the mean mSEPT9 level, and there was a good correlation between them.
We further studied the relationship between the plasma mSEPT9 level and the sensitivity of various types of adenoma. It can be seen from Figure 1C and 1D that significant increase in mSEPT9 level was not seen until TVA, and the mSEPT9 level kept increasing in VA and stage I CRC. Good correlation between the sensitivity and mSEPT9 level can also be observed in Figure 1D. It appeared that the villous component in adenoma is one crucial factor that led to the increase in mSEPT9 level and sensitivity.
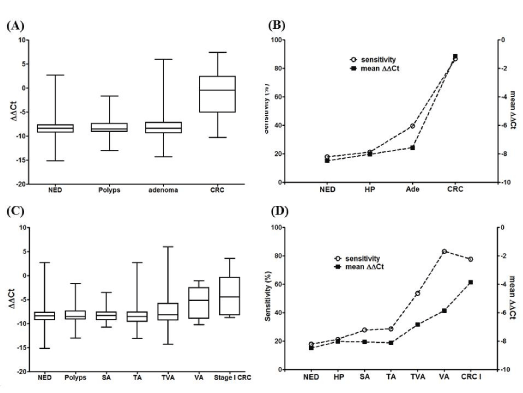
Figure 1. The methylation level (ΔΔCt) and the sensitivity of mSEPT9 in various colorectal diseases. The plasma mSEPT9 level for normal subjects (NED), subjects with HP, adenoma or CRC is shown in panel A as the box and whisker plot, and the mean values of mSEPT9 level and the detection sensitivity was compared in panel B. Similarly, the plasma mSEPT9 level for SA, TA, TVA and VA was shown in panel C as the box and whisker plot, and the mean values of mSEPT9 level and the detection sensitivity was compared in panel D. Data from NED, HP and Stage I CRC groups was also shown in panel C and D for comparison.
Plasma mSEPT9 level correlated with the degree of dysplasia in adenoma and determined the sensitivity
In order to further understand the relationship between the plasma mSEPT9 level and the pathological changes in adenoma, we investigated the mSEPT9 level and the corresponding sensitivity in adenomas without dysplasia (ND), adenomas with low-grade dysplasia (LGD) and adenomas with high-grade dysplasia (HGD). It can be clearly observed in Figure 2 that in SA (A), TA (B), TVA (C) and VA (D), adenomas with LGD generally exhibited higher mSEPT9 level and higher sensitivity than those without dysplasia, and adenomas with HGD generally exhibited higher mSEPT9 level and higher sensitivity than those with LGD. The mSEPT9 level exhibited perfect correlation with the detection sensitivity in all four types of adenoma. It can be suggested from Figure 2 that the degree of dysplasia is a decisive factor for the plasma mSEPT9 level and the sensitivity.
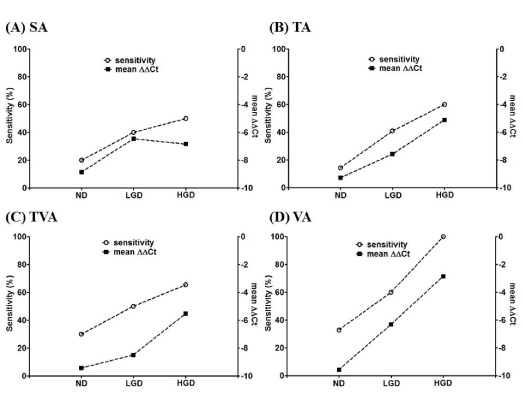
Figure 2. The plasma mSEPT9 levels (ΔΔCt) and the detection sensitivity of adenomas with various degrees of dysplasia. The plasma mSEPT9 level and the corresponding sensitivity for no dysplasia (ND), low-grade dysplasia (LGD) and high-grade dysplasia (HGD) for SA (panel A), TA (panel B), TVA (panel C) and VA (panel D) were presented and compared.
We summarized the data in this study in Figure 3 by plotting the sensitivity in all groups of subjects against the mSEPT9 levels in the corresponding groups. The data from all groups scatters in a wide range due to the wide distribution of mSEPT9 level and sensitivity. However, a clear linear relationship can be observed between the two. The relationship can be fit with a linear equation y=9.196x+109.7 with the r2 at 0.804, indicating the goodness of fit. The linear relationship shown in Figure 3 clearly suggests that the detection sensitivity is determined by the plasma mSEPT9 level.
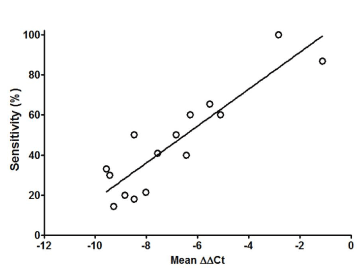
Figure 3. Linear regression on the relationship between detection sensitivity and plasma mSEPT9 level (ΔΔCt). Data presented in this figure was from NED, HP, adenomas (including ND, LGD and HGD for SA, TA, TVA and VA) and CRC investigated in this study, and was used to perform the regression.
The relationship between detection sensitivity and plasma mSEPT9 level
The data in this study clear demonstrated the correlation between the mSEPT9 level and the detection sensitivity and identified a linear relationship between the mSEPT9 level (ΔΔCt) and the sensitivity. With specific focus on various pathological types of adenoma, we proved that villous component in adenoma is one crucial factor leading to higher plasma mSEPT9 level and higher detection sensitivity. This is also true for the degree of dysplasia, in which adenoma with HGD exhibited higher plasma mSEPT9 level and detection sensitivity than those with LGD and without dysplasia.
The plasma mSEPT9 level differs from person to person in all groups investigated in this study, and this is why the mSEPT9 level distributed in a wide range, however, patients with more severe diseases generally exhibited higher mSEPT9 level than those with less severe diseases. Although this may not be true for each individual, the trend is clear if population is compared. It is not known why the mSEPT9 level of individuals in one group exhibited such a big variation, but several factors may affect the mSEPT9 level in plasma. First, since the circulating tumor DNA (ctDNA) in human blood exhibits dynamic balance and its half-life is quite short, different metabolism and in vivo environment of each individual may affect the ctDNA level. Secondly, sampling time affects the plasma mSEPT9 level in each individual, and it appeared that blood samples collected early in the morning exhibited the highest mSEPT9 level [11,12]. Thirdly, the growth pattern of tumor in each individual may varies, which leads to differential release of mSEPT9 DNA into circulation.
The relationship between disease severity and plasma mSEPT9 level
It can be suggested that the plasma level of mSEPT9 is closely correlated with the pathological lesions, and more severe or advanced lesions lead to higher level of mSEPT9 level and higher sensitivity. This observation further suggests that more advanced lesions may experience higher level of cell turnover and therefore release more DNA into circulation. As the colorectal lesions grow bigger and become more invasive, more DNA is released and the chance to detect it by the mSEPT9 assay also becomes higher. Although we focused on the relationship between mSEPT9 level and detection sensitivity in adenoma in this study, we found that the above trend is also true for CRC, in which patients with later stage CRC generally exhibited higher level of mSEPT9 and higher sensitivity than those with early stage CRC. This is reflected in previous observations using the SEPT9 assay for CRC detection [4].
Potential applications of quantitative mSEPT9 measurement in CRC diagnosis and therapy
As clear trend in quantitative mSEPT9 level was observed in various types of colorectal diseases, it appeared that the mSEPT9 level may be used as a scale for severity of diseases and therefore can potentially monitor the progression of diseases or therapeutic effect flowing surgery or radio- or chemotherapy. However, as the mSEPT9 level in each individual varies greatly, the levels can only be compared in an individual and cannot be compared between individuals. Technically, the effects of factors that may cause the mSEPT9 level fluctuation should be minimized to ensure the fidelity of comparison. Furthermore, comparisons can only be performed if the mSEPT9 levels fall into the linear scope of the qPCR standard curve, as values outside the linear scope cannot be accurately quantified, and qualitative interpretation should be used in this situation. The use of the mSEPT9 assay for CRC therapeutic effect assessment has a promising perspective and is currently under investigation.
In conclusion, linear correlation was found between the detection sensitivity and the level of plasma mSEPT9, which was positively correlated with the pathological severity. Adenomas with villous components or HGD exhibited higher mSEPT9 level and higher sensitivity than those without villous components or HGD. Our study suggests that the detection sensitivity of the mSEPT9 assay in colorectal adenoma was determined by the plasma level of mSEPT9, which reflects the severity of lesions.
Dr. Lele Song was an employee of BioChain (Beijing) Science and Technology, Inc. BioChain is a collaborator of Epigenomics AG, a Germany-based company that launched the first commercial SEPT9 assay.
This study was supported by the ‘clinical research support fund of the Chinese PLA General Hospital’ under the project named ‘the screening performance of colorectal adenoma by the methylated Septin9 assay alone or in combination with the fecal immunochemical test’.
- Schreuders EH, Ruco A, Rabeneck L, Schoen RE, Sung JJ, et al. (2015) Colorectal cancer screening: a global overview of existing programmes. Gut 64: 1637-1649. [Crossref]
- Church TR, Wandell M, Lofton-Day C, Mongin SJ, Burger M, et al. (2014) PRESEPT Clinical Study Steering Committee, Investigators and Study Team: Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut 63: 317-325.
- Potter NT, Hurban P, White MN, Whitlock KD, Lofton-Day CE, et al. (2014) Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin Chem 60:1183-1191. [Crossref]
- Song L, Jia J, Peng X (2017) The performance of the SEPT9 gene methylation assay and a comparison with other CRC screening tests: A meta-analysis. Sci Rep 7: 3032. [Crossref]
- Grützmann R, Molnar B, Pilarsky C, Habermann JK, Schlag PM, et al. (2008) Sensitive detection of colorectal cancer in peripheral blood by septin 9 DNA methylation assay. PLoS One 3: e3759. [Crossref]
- Tanzer M, Balluff B, Distler J, Hale K, Leodolter A, et al. (2010) Performance of epigenetic markers SEPT9 and ALX4 in plasma for detection of colorectal precancerous lesions. PLoS One 5: e9061. [Crossref]
- Warren JD, Xiong W, Bunker AM, Vaughn CP, Furtado LV, et al. (2011) Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med 9: 133. [Crossref]
- Jin P, Kang Q, Wang X, Yang L, Yu Y, et al. (2015) Performance of a second-generation methylated SEPT9 test in detecting colorectal neoplasm. J Gastroenterol Hepatol 30: 830-833. [Crossref]
- Song L, Peng X, Li Y, Xiao W, Jia J, et al. (2017) The SEPT9 gene methylation assay is capable of detecting colorectal adenoma in opportunistic screening. Epigenomics 9: 599-610. [Crossref]
- Dietrich D, Hasinger O, Liebenberg V, Field JK, Kristiansen G, et al. (2012) DNA methylation of the homeobox genes PITX2 and SHOX2 predicts outcome in non-small-cell lung cancer patients. Diagn Mol Pathol 21: 93-104. [Crossref]
- Yong E (2014) Cancer biomarkers: Written in blood. Nature 511: 524-526. [Crossref]
- Tóth K, Patai AV, Kalmár A, Barták BK, Nagy ZB, et al. (2017) Circadian Rhythm of Methylated Septin 9, Cell-Free DNA Amount and Tumor Markers in Colorectal Cancer Patients. Pathol Oncol Res 23: 699-706. [Crossref]



