Reaction of organotin dichlorides with the salt of 2-ketoglutaric acid is rapid forming organotin polyether polymers in low to moderate yield within 15 seconds or less employing the interfacial polycondensation process. The average chain length generally decreases as the alkyl chain length on the organotin increases. Infrared spectroscopy shows bands characteristic of both reactants and the formation of the linkage ester linkage. MALDI MS shows formation of ion fragments characteristic of 5 to 8 repeat units and good isotopic abundance matches consistent with the presence of the organotin moiety in these ion fragments. NMR is also consistent with the formation of the organotin polyester. The polymers exhibit good inhibition of all cancer cell lines tested including two breast and two pancreatic cancer cell lines.
cancer, 2-ketoglutaric acid, organotin polymers, pancreatic cancer, breast cancer
This paper is part of an ongoing effort to synthesize metal-containing condensation polymers that can be used to treat various biological problems. Typically, we employ a metal-containing Lewis acid, here an organotin dihalide, reacted with a biologically active Lewis base. This work has been recently reviewed [1].
2-Ketoglutaraic acid, KA, is a key member of the Krebs cycle. Its anion is called alpha-ketoglutarate. It is an essential nitrogen transporter in metabolic pathways. The amino groups of amino acids are connected to 2-ketoglutarate through amide formation and eventually transported to the liver where the urea cycle occurs. 2-Ketoglutarate is aminated, along with glutamine, forming the neutrotransmitter glutamate which can be decarboxylated forming the inhibitory neutrotransmitter GABA. It also plays an essential role in the detoxification of ammonia in the brain [2]. 2-Ketoglutaraic acid also plays an important role in redox reactions involving molecular oxygen. Molecular oxygen oxidizes many biological compounds producing important compounds including antibiotics in reactions catalyzed by oxygenases. Along with oxygen, 2-ketoglutarate is required for the hydroxylation of proline to hydroxyproline in the synthesis of Type 1 collagen [3,4].
2-Ketoglutarate has recently been implicated in other important biological behaviors. For instance, it has been implicated in increasing the lifetime of nematode worms [5]. It has also been suggested that it is involved in immune regulation [6]. Thus it has been related to having numerous biological activities.
There has not been reported the prior synthesis of condensation polymers directly from 2-ketoglutaric acid. But there are reports of its inclusion of polymers. There have been several reports on the modification of chitosan trough reaction between the chitosan and KA forming a linkage between the amine on the chitosan and methylene group on the KA [7-9]. This product is further reacted with an aqueous solution of iron III chloride forming chelated products that release KA in a sustained-release fashion.
Barrett and Yousal reported the synthesis of elastomers based on the thermal polycondensation of KA and triols [10,11]. The products undergo hydrolysis degradation releasing KA.
There are several reports involving the chelating of KA with various metal ions. For instance, Deng and co-workers reported the synthesis of MOF products though chelation with the copper ion [12]. Ferari and co-workers reported similar properties also from chelation with the copper ion [13].
Here we describe the incorporation of 2-ketoglutaric acid into organotin polyester polymers from reaction with various organotin dihalide forming general repeat units as shown below (Figure 1).
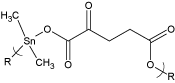
Figure 1. Repeat units for the product of dimethyltin dichloride and the salt of 2-ketoglutaric acid where R represents simple chain extension.
The actual form of the 2-ketoglutaric acid in the reaction is the disalt since the non-salt form is not a sufficiently strong Lewis base to form an ester linkage (Figure 2).
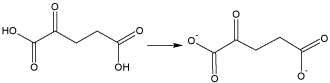
Figure 2. Structure of the active Lewis base, the disalt of 2-ketoglutaric acid. The structural characterization and preliminary anticancer activity is also given.
Synthesis
Reactions were carried out using the interfacial polycondensation technique. Briefly, an aqueous solution (30 ml) containing the KA (0.00300 mol) and sodium hydroxide (0.0060 mol) was transferred to a one quart Kimax emulsifying jar fitted on top of a Waring Blender (model 1120; no load speed of about 18,000 rpm; reactions were carried out at about 25 oC). Stirring was begun and a hexane solution (30 ml) containing the organotin dihalide (0.00300 mol) was rapidly added (about 3-4 seconds) through a hole in the jar lid using a powder funnel. The resulting solution was blended for 15 seconds. The precipitate was recovered using vacuum filtration and washed several times with deionized water and hexane to remove unreacted materials and unwanted by-products. The white solid was washed onto a glass petri dish and allowed to dry at room temperature.
Diphenyltin dichloride (1135-99-5), 2-ketoglutaric acid (328-50-7) and dibutyltin dichloride (683-18-1) were purchased from Aldrich Chemical Co., Milwaukee, WS; diethyltin dichloride (866-55-7) was obtained from Peninsular Chemical Res., Gainesville, FL; dioctyltin dichloride (3542-36-7), was obtained from Ventron Alfa Inorganics, Beverly, Mass.
Physical Characterization
Molecular weight was determined employing light scattering photometry. Light scattering photometry was carried out employing a Brice-Phoenix Universal Light Scattering Photometer Model 4000. Infrared spectra were obtained employing attenuated total reflectance infrared spectroscopy utilizing a Thermo Scientific Nicolet iS5 FTIR equipped with an id5 ATR attachment. 1H NMR spectra were obtained employing Varian Inova 400 MHz and Varian 500 MHz spectrometers.
High resolution electron impact positive ion matrix assisted laser desorption ionization time of flight, HR MALDI-TOF, mass spectrometry was carried out employing a Voyager-DE STR BioSpectrometer, Applied Biosystems, Foster City, CA. The standard settings were used with a linear mode of operation and an accelerating voltage of 25,000 volts; grid voltage 90% and an acquisition mass range of 500 to 2,500 Da. A graphite matrix was employed. Graphite from a number 2 pencil was marked on the sample holder and sample placed onto the graphite mark.
The toxicity of each test compound was evaluated with the human pancreas adenocarcinoma cell line (AsPC-1), human pancreas epithelioid duct carcinoma cell line (PANC-1) or other cell line. Following a 24 h incubation period, the test compounds were added at concentrations ranging from 0.0032 to 32,000 ng/mL and allowed to incubate at 37°C with 5% CO2 for 72 h. Following incubation, Cell Titer-Blue reagent (Promega Corporation) was added (20 µL/well) and incubated for 2 h. Fluorescence was determined at 530/590 nm and converted to % cell viability versus control cells.
All cytotoxicity values are calculated against a base-line value for each line that was generated from “mock-treatment” of the normal and tumor cell lines with media supplemented with all diluents used to prepare the chemotherapeutic compounds. For example, if the compounds were dissolved in DMSO and serial dilutions prepared in MEM to treat the cells, then the mock-treated cells were “treated” with the same serial dilutions of DMSO without added chemotherapeutic compound. This was done to ensure that any cytotoxicity observed was due to the activity of the compound and not the diluents. For the studies reported here, the mock-treatment never resulted in a loss of cell viability of more than one percent, demonstrating that the activity observed was not due to cytotoxicity of any of the diluents used, but was due to activity of the tested compounds.
Yields and Chain Lengths
Product yield and chain length is given in Table 1. As previously noted the reaction involves the disalt of 2-ketoglutaric acid rather than simply 2-ketoglutaric acid itself since the acid is not a sufficient nucleophile to displace the chloride on the organotin dichloride whereas the salt readily displaces the halide forming the ester linkage. The linkage is referred to as an ester linkage because in the naming of these materials, the organometallic is named as a methylene-like moiety or unit. Thus, the created linkage is an ester.
Table 1. Product yield and chain length for the synthesis of organotin poly(ester ethers) from reaction of organotin dihalides with 2-ketoglutaric acid.
Organotin Moiety |
Percentage Yield |
Molecular Weight |
Chain Length |
Me2Sn |
16 |
9.6 x 104 |
320 |
Et2Sn |
37 |
1.4 x 105 |
430 |
Bu2Sn |
46 |
1.9 x 105 |
500 |
Oc2Sn |
4 |
1.7 x 105 |
340 |
Ph2Sn |
90 |
6.7 x 105 |
160 |
In general there is an increase in yield and chain length as the alkyl group increases in length from methyltin dichloride to the butyltin dichloride. Solubility in hexane increases as the alkyl group is larger favoring higher yields for the smaller alkyltins since they are forced from the organic layer towards the salt of the ketoglutaric acid. It is possible that the lower solubility of the smaller alkyltins prevent ready reaction with the salts of 2-ketoglutaric acid and this is responsible for the observed trend. It must be noted that solubility of the salt of ketoglutaric acid in the organic layer, here heptane, is low possibility forcing reaction to occur within the aqueous layer. Low yield for dioctyltin is typical and may be due to the bulkiness of the octyl group inhibiting easy availability to the metal atom for reaction.
Infrared Results
Infrared spectral analysis was carried out for all of the samples over the range of 4000-400 cm-1. All band locations are given cm-1.
Infrared spectral analysis is consistent with the proposed structure and with other reported analyses for KA [14-16] and organotin polymers [17-19]. Table 2 contains results for the dibutyltin and diphenyltin polymers. All spectra show bands characteristic of both reactants and new bands for the products assigned to the Sn-O linkage
Table 2. Assigned peaks for the monomers and associated polymers derived from reaction with 2-ketoglutaric acid and dibutyltin dichloride and diphenyltin dichloride.
Band Assignment |
2-Ketoglutaric Acid, KA |
Bu2SnCl2 |
Bu2Sn/ Polymer |
Ph2SnCl2 |
Ph2Sn/ Polymer |
CH aromatic |
|
|
|
3068,3051 |
3068,3047 |
CH st |
2941,2929 |
|
2940,2930 |
|
2941,2930 |
CH3 asym st |
|
2959 |
2957 |
|
|
CH2 asym st |
|
2926 |
2926 |
|
|
CH3 sym st |
|
2872 |
2874 |
|
|
CH2 sym st |
|
2858 |
2857 |
|
|
C=O, internal |
1723 |
|
1734 |
|
1737 |
C=O |
1690,1640 |
|
1599,1590 |
|
1617,1600 |
δ CH2 |
1442,1408 |
|
1462,1412 |
|
1479,1430 |
ω CH2 |
1329 |
|
1341 |
|
1332 |
C-C st |
1093,1047 |
|
1109,1049 |
|
1101,1068 |
Sn-Ph st |
|
|
|
1480 |
1480 |
C=C st |
|
|
|
1432 |
1431 |
Sn-O asy st |
|
|
1290 |
|
1287 |
Sn-Ph st |
|
|
|
1071 |
1068 |
C-C st |
|
1178,1152 |
1180,1158 |
|
|
C-O st |
1226 |
|
1120 |
|
1126 |
Sn-O(CO)st |
|
|
1025 |
|
1021 |
Ring breathing |
|
|
|
996 |
997 |
CH3 rock |
|
878 |
880 |
|
|
Sn-O sym st |
|
|
775 |
|
761 |
Sym op bend H’s |
|
|
|
729 |
727 |
Asy op bend ring |
|
|
|
691 |
691 |
Sn-CH2 asym st |
|
592 |
597 |
|
|
Sn-CH2 sym st |
|
509 |
509 |
|
|
OH op wag |
577 |
|
|
|
|
Sn-Ph asym st |
|
|
|
442 |
440 |
There is only minor change in the location of the internal carbonyl but there is a major difference in the external of carboxylic carbonyl positions as expected since there is a major difference in the environment of this carboxyl environment.
Bands consistent with the formation of the Sn-O bond are found corresponding to the symmetric and asymmetric stretches and a band is found assigned to the stretching for the Sn connected to the oxygen of the carboxylic moiety. Table 3 contains band locations for each of the synthesized polymers.
Table 3. Assigned peaks associated with the formation of the Sn-O linkage.
Organotin Moiety |
Sn-O Asym. St. |
Sn-O-(CO) St. |
Sn-O Sym. St. |
Me2Sn |
1278 |
1029 |
787 |
Et2Sn |
1278 |
1020 |
759 |
Bu2Sn |
1290 |
1025 |
772 |
Oc2Sn |
1279 |
1022 |
768 |
Ph2Sn |
1287 |
1025 |
761 |
The polyesters can exist as bridging or distorted octahedral and non-bridging or distorted tetrahedral about the tin (Figure 3) [1]. Infrared spectroscopy is the easiest way to determine the structure about the tin. Bridging asymmetric carbonyl absorptions are found around 1540-1580 (all infrared bands are given in cm-1). The bridging symmetric carbonyl band is found around 1400-1435. Non-bridging asymmetric carbonyl bands are found about 1590-1650; and the corresponding symmetric carbonyl band found about 1350-1390.
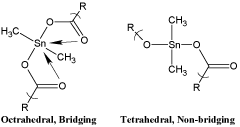
Figure 3. Bridging and non-bridging structures about the tin atom for the product derived from dimethyltin dichloride.
Table 4 contains results for all of the products with respect to bands associated to bridging/non-bridging. The organotins containing the smallest alkyl groups favor the non-bridged structures while those with larger alkyl groups and the phenyl group are a mix consisting of both bridged and non-bridges geometries about the tin atom. The bridged structure is called an octahedral geometry about the tin because geometrically it forms two tetrahedral structures each with four faces.
Table 4. Presence of bridging and non-bridging associated bands and location.
Organotin Moiety |
Asym. Nonbridging |
Sym Nonbridging |
Asym Bridging |
Sym Bridging |
Me2Sn |
1615(l) |
1380(l) |
- |
- |
Et2Sn |
1605(l) |
1379(l) |
- |
- |
Bu2Sn |
1599(l),1590(l) |
1377(l),1370(l) |
1572(m) |
1412(m) |
Oc2Sn |
1616(l),1608(l) |
1385(l),1376(l) |
1562(m) |
1407(m) |
Ph2Sn |
1617(s),1600(s) |
1388(m),1380(m) |
1577(m),1561(m) |
1431(l) |
MALDI MS
We have been investigating the solid-state fragmentation of various polymers employing MALDI MS emphasizing metal-containing polymers for use in the structural identification of these polymers. Matrix-assisted desorption/ionization mass spectrometry was independently introduced in 1981 by Barber and Liu and coworkers [20,21]. The addition of the laser as the energy source was introduced by Tanaka, Hillenkamp and coworkers in 1988 [22,23]. The combinations of these concepts allowed the creation of matrix-assisted laser/desorption mass spectroscopy, MALDI MS. We have been using a modification of this technique that allows MALDI MS to be obtained on non-volatile and insoluble products. This approach has been recently reviewed [24-26].
MALDI MS spectra were obtained for the polymers. Recently we have been employing graphite as the matrix material because it gives good results with few interfering ion fragments produced above 500 mass which is the typical lower mass range employed in our studies [27,28]. Two general MALDI MS modes were employed. These are the reflective and linear modes. The reflective mode has a longer focal length than the linear mode. Results for the reflective mode allow finer features, such as isotopic abundances, to be more accurately determined but generally results in the detection of lower masses. By comparison, the linear mode has a shorter flight distance and results in the detection of higher masses. Following are results for two of the polymers.
A portion of the MALDI MS for the dioctyltin polymer is given in Figures 4 and 5. Each of the ion fragment clusters above 500 (all ions are given in daltons, Da, or m/e =1) are actually clusters of ions that are produced because of the presence of tin atom(s) within each cluster. Because tin has isotopes, different ion fragments are created that have the same structural formula but vary by the particular tin isotope present. This creates what is often referred to as spectral “fingerprints” characteristic of the natural abundance of these isotopes. The fragments given in the following tables are actually clusters of such ion fragments. Along with the pictorial representations as given in Figures 4 and 5, such distributions can also be presented in table form and compared with known values. Table 5 contains the most abundant ion fragment clusters from Figures 4 and 5. The following abbreviations are utilized to describe the tentative ion fragment cluster assignments- KA = 2-ketoglutaric acid minus two hydrogen atoms; Oc2Sn = dioctyltin moiety, O = oxygen, U = one repeat unit; 2U = two repeat units; Etc. Additional less abundant ion fragment clusters are also present. Sodium is a common contaminant.
Figure 4. MALDI MS for the product of dioctyltin dichloride and the salt of 2-ketoglutaric acid using the reflective mode with graphite matrix over the approximate range of 500 to 2500 Da.
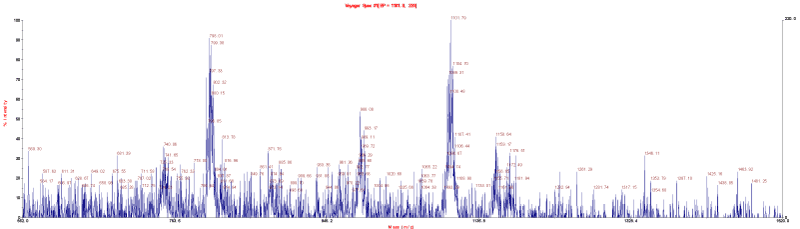
Figure 5. MALDI MS for the product of dioctyltin dichloride and the salt of 2-ketoglutaric acid using the linear mode with graphite matrix over the approximate range of 500 to 2500 Da.
Table 5. Most abundant ion fragment clusters for the product of dioctyltin dichloride and 2-ketoglutaric acid over the approximate mass range of 500 to 2500 Da.
Mass,Da/ Linear |
Mass,Da/Reflective |
(Tentative) Assignment |
569 |
|
U+2CO2,Na |
740 |
740 |
2U+OcSnO |
798 |
801 |
U+Oc2Sn,CO2,Na |
|
875 |
U+ Oc2Sn,2O |
|
901 |
U+ Oc2Sn,CO2,Na |
989 |
|
2U |
1101 |
1102 |
2U+KA-O |
1150 |
|
2U+KA,Na |
1261 |
|
2U+OcSn,O |
|
1278 |
2U+OcSn,CO2,Na |
1346 |
|
2U+ Oc2Sn,O |
|
1360 |
2U+ Oc2Sn,2O |
|
1401 |
2U+ Oc2Sn,CO2,Na |
1463 |
|
3U |
1549 |
|
3U+CO2,Na |
|
1599 |
3U+KA-2O |
1645 |
|
3U+KA,Na |
1707 |
|
3U+OcSn |
1802 |
|
3U+OcSn,2CO2 |
|
1816 |
3U+ Oc2Sn |
1887 |
|
3U+ Oc2Sn,CO2,O |
|
2011 |
4U+CO2 |
2082 |
|
4U+KA-2O |
2200 |
|
4U+OcSn |
|
2260 |
4U+ OcSn,CO2,Na |
2316 |
|
5U+ Oc2Sn |
|
2429 |
5U+ Oc2Sn,2CO2,Na |
Ion fragments to five units are found. Loss of the organic substitutes on the metal atom can occur and appear only at the site of chain scission [24-28]. This is found here.
As noted before, isotopic abundance matches can be employed to support the presence of tin atoms in the isotopic fragment clusters. Tin contains ten isotopes of which seven are considered significant (a natural abundance >5%). At higher masses, isotope matches are difficult because of the low intensities of generated ion fragments. Even so, at lower masses such isotope matches are possible. Table 6 contains two matches for ion fragment clusters containing two tin atoms. The abundance matches are consistent with the presence of two tin atoms within the particular ion fragment clusters.
Table 6. Isotopic abundance matches for two tin-containing ion fragment clusters containing two tin atoms (Only ion fragments >5% relative abundance are reported.) for the product of dioctyltin dichloride and 2-ketoglutaric acid.
Known for Sn |
2U |
2U+K-O |
Da |
% Rel Abu |
Da |
% Rel Abu |
Da |
% Rel Abu |
232 |
12 |
983 |
14 |
1096 |
12 |
233 |
13 |
984 |
15 |
1097 |
14 |
234 |
43 |
985 |
42 |
1098 |
40 |
235 |
35 |
986 |
34 |
1099 |
36 |
236 |
94 |
987 |
94 |
1100 |
96 |
237 |
51 |
988 |
53 |
1101 |
51 |
238 |
100 |
989 |
100 |
1102 |
100 |
239 |
35 |
990 |
37 |
1103 |
35 |
240 |
81 |
991 |
82 |
1104 |
82 |
242 |
32 |
993 |
30 |
1106 |
34 |
244 |
22 |
995 |
24 |
1108 |
24 |
The “mildness” of MALDI MS is shown by the lack of fragmentation of the C(O)CH2CH2 chain.
Table 7 contains the most abundant ion fragments for the product of dibutyltin dichloride and 2-ketoglutaric acid over the approximate range of 500 to 2500 Da.
Table 7. Most abundant ion fragment clusters for the product of dibutyltin dichloride and 2-ketoglutaric acid over the approximate mass range of 500 to 2500 Da.
Mass,Da/
Linear |
Mass,Da/
Reflective |
(Tentative) Assignment |
|
523 |
U+KA |
586 |
588 |
U+BuSn,O,Na |
|
615 |
U+Bu2Sn |
653 |
655 |
U+Bu2SnCO2 |
671 |
672 |
U+ Bu2Sn,CO2,O |
703 |
705 |
U+ Bu2Sn,2CO2 |
740 |
742 |
2U-O |
|
755 |
2U |
766 |
766 |
2U+Na-O |
840 |
839 |
2U+KA-CO2,O |
857 |
860 |
2U+KA-CO2 |
938 |
938 |
2U+BuSn |
1024 |
|
2U+ Bu2Sn,2O |
1061 |
|
2U+ Bu2Sn,CO2,Na |
1096 |
|
2U+ Bu2Sn,2CO2,Na |
|
1148 |
3U |
1266 |
|
3U+KA-O |
1284 |
|
3U+KA |
1367 |
|
3U+ Bu2Sn |
1389 |
|
3U+ Bu2Sn,Na |
|
1420 |
3U+ Bu2Sn,CO2 |
|
1535 |
4U+Na |
1558 |
|
4U+CO2 |
1641 |
1639 |
4U+KA-O |
1658 |
1658 |
4U+KA |
1751 |
|
4U+ Bu2Sn |
|
1765 |
4U+ Bu2Sn,O |
1852 |
|
5U-CO2 |
1953 |
|
5U+CO2,Na |
|
2056 |
5U+KA,Na |
2153 |
|
5U+ Bu2Sn,2O |
|
2161 |
5U+ Bu2Sn,O,Na |
2214 |
|
5U+ Bu2Sn,2CO2 |
2271 |
|
6U |
|
2300 |
6U+Na |
|
2392 |
6U+KA-O |
Ion fragments to six units are found. Again, the mildness of the MALDI MS system is demonstrated by lack of bond scission for the C(O)CH2CH2 moiety.
Table 8 contains an isotopic abundance match for one ion fragment cluster containing one tin atom. Table 9 contains a similar isotopic abundance match for a fragment cluster containing two tin atoms. Both show reasonable matches between what is found and what is expected consistent with the presence of one and two tin atoms in the corresponding ion clusters.
Table 8. Isotopic abundance match for an ion fragment cluster containing one tin atom derived from the product of dibutyltin dichloride and 2-ketoglutaric acid (only ion fragments >5% relative abundance are reported).
Known for Sn |
U+K |
Da |
% Rel Abu |
Da |
% Rel Abu |
116 |
45 |
519 |
46 |
117 |
24 |
520 |
23 |
118 |
75 |
521 |
76 |
119 |
26 |
522 |
25 |
120 |
100 |
523 |
100 |
122 |
14 |
525 |
16 |
124 |
17 |
527 |
19 |
Table 9. Isotopic abundance match for an ion fragment cluster containing two tin atoms derived from the product of dibutyltin dichloride and 2-ketoglutaric acid (only ion fragments >5% relative abundance are reported).
Standard/Known for 2 Sn |
U+Bu2Sn,CO2 |
Da |
% Rel Abu |
Da |
% Rel Abu |
232 |
12 |
649 |
11 |
233 |
13 |
650 |
12 |
234 |
43 |
651 |
43 |
235 |
35 |
652 |
34 |
236 |
94 |
653 |
90 |
237 |
51 |
654 |
47 |
238 |
100 |
655 |
100 |
239 |
35 |
656 |
31 |
240 |
81 |
657 |
85 |
242 |
32 |
659 |
35 |
244 |
22 |
661 |
23 |
Table 10 contains the major ion fragment clusters for the product of dimethyltin dichloride and 2-ketoglutaric acid. Ion fragment clusters to eight units are found. Again, the C(O)CH2CH2 unit remains intact.
Table 10. Most abundant ion fragment clusters for the product of dimethyltin dichloride and 2-ketoglutaric acid over the approximate mass range of 500 to 2500 Da.
Mass,Da/
Linear |
Mass,Da/
Reflective |
(Tentative) Assignment |
577 |
573 |
2U-O |
623 |
|
2U+CO2 |
|
688 |
2U+KA-CO2 |
694 |
696 |
2U+KA-2O |
770 |
767 |
2U+Me2Sn,2O |
797 |
801 |
2U+ Me2Sn,CO2,O |
874 |
878 |
3U |
919 |
|
3U+O,Na |
969 |
|
3U+KA-CO2,O |
|
1044 |
3U+ Me2Sn,O |
1087 |
|
3U+ Me2Sn,2O,Na |
1175 |
1173 |
4U |
|
1206 |
4U+O,Na |
1231 |
1233 |
4U+KA-2CO2 |
|
1333 |
4U+ Me2Sn |
1406 |
1408 |
4U+ Me2Sn,CO2,O |
1563 |
|
5U+KA-CO2 |
|
1585 |
5U+KA-2O |
1608 |
|
5U+KA |
|
1674 |
5U+ Me2Sn,2O,Na |
|
1738 |
6U+Na-CO2 |
1785 |
|
6U+Na |
1845 |
1846 |
6U+KA-CO2,O |
|
1888 |
6U+KA-O |
1954 |
|
6U+ Me2Sn,O,Na |
2007 |
|
7U-CO2 |
2071 |
|
7U+O |
2158 |
|
7U+KA-CO2 |
|
2223 |
7U+ Me2Sn,O |
2318 |
2320 |
8U-CO |
|
2402 |
8U-2CO2 |
In all cases the major bond scission occurs at the heteroatom sites within the polymer chain as shown in Figure 6. This is what has been found for other similar organotin polymers [1].
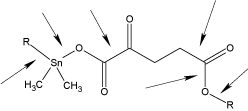
Figure 6. Preferred bond locations for chain scission.
Proton NMR
Proton NMR was carried out on the monomers and polymers in d-6 DMSO. All bands are given ppm. KA shows two bands at 2.4 and 3.1 assigned to the methylene hydrogens and about 13 for the hydroxyl protons which is absent in the polymers [29,30]. Dioctyltin shows bands from about 2.1 (nearest to the tin) to multiple bands about 1.2 furthest from the tin. The dioctyltin polymer shows bands at 2.5 and 3.2 from KA and bands at 2.1 and multiple bands about 1.3 to 1.2. Diphenyltin dichloride shows bands at 7.9 (ortho), 7.4 and 7.3 (meta and para). The corresponding polymer exhibits bands at 8.1, 7.7 and 7.6 from the diphenyltin moiety and 2.48 and 3.1 from KA.
Thus, proton nmr shows bands derived from both reactants within the products. There are only mild shifts between the monomers and polymer. Because of the poor solubility of the polymer, additional data is not confidently available.
Cell Analysis Results
Cell lines employed in the current study are given in Table 11.
Table 11. Cell lines employed in the current study.
Strain # |
NCI Desig. |
Species |
Tumor Origin |
Histological Type |
3465 |
PC-3 |
Human |
Prostate |
Carcinoma |
7233 |
MDA MB-231 |
Human |
Pleural effusion breast |
Adenocarcinoma |
1507 |
HT-29 |
Human |
Recto-sigmoid colon |
Adenocarcinoma |
7259 |
MCF-7 |
Human |
Pleural effusion-breast |
Adenocarcinoma |
ATCC CCL-75 |
WI-38 |
Human |
Normal embryonic lung |
Fibroblast |
CRL-1658 |
NIH/3T3 |
Mouse |
Embyro-continuous cell line of highly
contact-inhibited cells |
Fibroblast |
|
AsPC-1 |
Human |
Pancreatic cells |
Adenocarcinoma |
|
PANC-1 |
Human |
Epithelioid pancreatic cells |
Carcinoma |
The cells represent a broad range of cancers. The 3T3 cells are formally described as NIH 3T3 cells. Here, we will use both designations to describe this cell line.
Much of our recent effort has been on discovering compounds that inhibit pancreatic cancer because pancreatic cancer does not have a generally accepted "cure". Thus the set of tested cell lines includes two widely employed pancreatic cell lines. These are AsPC-1 which is an adenocarcinoma pancreatic cell line which represents about 80% of the observed cancers and PANC-1, which is an epithelioid carcinoma pancreatic cell line, representing about 10% of the observed human pancreatic cancers. The pair of breast cancer cell lines deserves special comment. They represent a matched pair of cell lines. The MDA-MB-231 (strain number 7233) cells are estrogen-independent, estrogen receptor negative while the MCF-7 (strain line 7259) cells are estrogen receptor (ER) positive. In some studies involving organotin polymers we found there was a marked difference between the ability to inhibit the two cell lines dependent on polymer structure [1]. The PC-3 (3465) cells are of interest because this particular prostate cell line is viewed as one of the most resistant of the prostate cancer cell lines.
While different measures have been employed in the evaluation of cell line results the most widely employed involves the concentration, dose, needed to reduce the growth of the particular cell line. Here we will use effective concentration, EC, values. The concentration of a drug, antibody, or toxicant that induces a response halfway between the baseline and maximum after a specified exposure time is referred to as the 50% response concentration and is given the symbol EC50.
Table 12 contains the EC50 values for the current polymers and monomers. Values for cisplatin are included. Cisplatin is among the most widely employed chemodrugs in the treatment of a wide variety of cancers.
Table 12. EC50 Concentrations (micrograms/mL) for the tested compounds. Values given in () are Standard Deviations for each set of measurements.
Sample |
3T3 |
WI-38 |
PANC-1 |
AsPC-1 |
Me2SnCl2 |
0.43 (.1) |
0.22(.1) |
0.80(.1) |
0.71(.1) |
Me2Sn/KA |
0.42(.04) |
0.44(.04) |
0.42(.04) |
0.41(.04) |
Et2SnCl2 |
0.46(.1) |
0.20(.1) |
0.48(.1) |
0.90(.1) |
Et2Sn/KA |
0.39(.04) |
0.39(.04) |
0.41(.04) |
0.38(.04) |
Bu2SnCl2 |
0.20 (.05) |
0.20(.05) |
0.0032(.001) |
0.012(.01) |
Bu2Sn/KA |
0.54(.05) |
0.52(.05) |
0.52(.06) |
0.55(.06) |
Oc2SnCl2 |
0.56(.1) |
0.30(.1) |
0.85(.1) |
0.85(.1) |
Oc2Sn/KA |
0.50(.04) |
0.49(.04) |
0.51(.04) |
0.45(.04) |
Ph2SnCl2 |
0.66(.1) |
0.25(.1) |
0.71(.1) |
0.83(.1) |
Ph2Sn/KA |
0.51(.05) |
0.45(.04) |
0.51(.04) |
0.50(.05) |
KA |
>32 |
>32 |
>32 |
>32 |
Cisplatin |
0.015(.01) |
0.019(.01) |
0.0023(.005) |
0.0035(.005) |
Sample |
PC-3 |
MDA-MB-231 |
HT-29 |
MCF-7 |
Me2SnCl2 |
0.51(.1) |
0.44(.1) |
0.56(.1) |
0.66(.1) |
Me2Sn/KA |
0.44(.4) |
0.45(.4) |
0.42(.5) |
0.46(.5) |
Et2SnCl2 |
0.61(.1) |
0.64(.1) |
0.71(.1) |
0.77(.1) |
Et2Sn/ KA |
0.38(.05) |
0.42(.1) |
0.45(.05) |
0.42(.05) |
Bu2SnCl2 |
1.4(1.1) |
1.4(1.3) |
1.2(.1) |
0.7(.06) |
Bu2Sn/ KA |
0.53(.06) |
0.51(.07) |
0.51(.08) |
0.54(.05) |
Oc2SnCl2 |
0.55(.1) |
0.65(.1) |
0.65(.1) |
0.70(.1) |
Oc2Sn/ KA |
0.49(.04) |
0.47(.06) |
0.51(.06) |
0.52(.06) |
Ph2SnCl2 |
0.82(.1) |
0.76(.1) |
0.56(.1) |
0.68(.1) |
Ph2Sn/ KG |
0.44(.04) |
0.48(.05) |
0.52(.05) |
0.51(.04) |
KG |
>32 |
>32 |
>32 |
>32 |
Cisplatin |
0.0044(.004) |
0.0029(.002) |
0.0041(.003) |
0.0057(.003) |
EC50 values are generally lower for the polymers compared with organotin monomers and KA exhibits no inhibition to the limit of testing. There does not appear to be a difference in the ability to inhibit growth for the two breast-associate cell lines. Also, the polymers exhibit decent inhibition of the two pancreatic cancer cell lines consistent with them possibly possessing an ability to inhibit other pancreatic cell lines. The polymers also exhibit decent inhibition of the cell lines of the other cancer-associated cells.
In other studies, we observed a marked ability for polymers containing the dibutyltin moiety, followed by those containing the diphenyltin moiety, to exhibit lower EC50 values. This is not the case here where there is similarity between the ability to inhibit cell growth as the organotin moiety varies [1]. This means that the polymer derived from dibutyltin dichloride can be employed to inhibit the cancer cell lines. This is advantageous since dibutyltin dichloride offers the least toxicity toward humans of the organotin compounds. Second, it is the least expensive of the employed organotin dihalides. Third, it is the most widely used of the organotin dihalides available in the gram to ton quantity. Finally, it decomposes to tin oxide which is environmentally considered non-toxic.
Another measure of the potential use of compounds is the concentration of drug necessary to inhibit the standard cells compared to the concentration of drug necessary to inhibit the growth of the test cell line. Again, a variety of symbols are employed to describe similar calculations. Here, we will simply employ the term chemotherapeutic index, CI, so that the CI50 is then the ratio of the EC50 for the standard cell line, NIH/3T3 or WI-38 cells, divided by the EC50 for the particular test cell.
Two cell lines are typically employed as standards in the evaluation of the effectiveness of compounds to arrest the growth of tumor cell lines. These two cell lines are the NIH/3T3 and WI-38 cell lines. We have begun comparing these two cell lines as biomarkers to study the effectiveness of compounds to inhibit the growth of various tumor cell lines.
NIH/3T3 cells are mouse embryo fibroblast cells. They are part of a group of cell lines that are referred to as partially transformed cells in that they are immortal unlike normal cells. They retain other characteristics of normal cells such as being contact-inhibited. Relative to most normal cells they are robust and easily maintained.
WI-38 cells are normal embryonic human lung fibroblast cells. They have a finite life time of about 50 replications. Compared to NIH/ 3T3 cells, they are more fragile and difficult to maintain for long periods of time. Thus, NIH/3T3 cells are often favored because of ease of handling aided by an infinite life span.
We have employed both cell lines in our studies and have only recently begun to compare results to see if there is a difference between the results found for the two cell lines when they are used to evaluate the ability of materials to arrest the growth of various cancer cell lines. Thus, this study has two parts embedded into it. One is the actual ability to control cancer and the second is the comparison of results derived from employing the two most common standard cells.
The CI50 values for the monomers and polymers are given in Table 13. For comparison, values for cisplatin are also given.
Table 13. CI50 results for values calculated from data given in Table 12.
Sample |
EC503T3/
EC50 WI-38 |
EC50 3T3/
EC50PANC-1 |
EC50 3T3/
EC50AsPC-1 |
EC50 3T3/
EC50PC-3 |
Me2SnCl2 |
2 |
0.54 |
0.61 |
0.84 |
Me2Sn/ KA |
0.95 |
1.0 |
0.1 |
0.95 |
Et2SnCl2 |
1.2 |
1.0 |
1.0 |
1.1 |
Et2Sn/ KA |
1.0 |
0.96 |
0.51 |
1.0 |
Bu2SnCl2 |
1.0 |
63 |
17 |
0.14 |
Bu2Sn/ KA |
1.1 |
1.0 |
0.99 |
0.74 |
Oc2SnCl2 |
1.9 |
0.66 |
0.66 |
1.0 |
Oc2Sn/ KA |
1.0 |
0.99 |
1.1 |
1.0 |
Ph2SnCl2 |
2.6 |
0.92 |
0.80 |
0.80 |
Ph2Sn/ KA |
1.1 |
1.0 |
1.0 |
1.2 |
Cisplatin |
0.80 |
6.5 |
4.3 |
3.8 |
Sample |
EC50 WI-38/
EC50PANC-1 |
EC50 WI-38/
EC50AsPC-1 |
EC50 WI-38/
EC50PC-3 |
EC50 WI-38/
EC50MDA |
Me2SnCl2 |
0.28 |
0.31 |
0.43 |
0.50 |
Me2Sn/ KA |
1.0 |
1.1 |
1.0 |
0.98 |
Et2SnCl2 |
0.83 |
0.81 |
0.91 |
0.91 |
Et2Sn/ KA |
0.98 |
1.0 |
1.0 |
0.93 |
Bu2SnCl2 |
63 |
17 |
0.14 |
0.14 |
Bu2Sn/ KA |
1.1 |
0.98 |
1.0 |
1.1 |
Oc2SnCl2 |
0.35 |
0.35 |
0.55 |
0.46 |
Oc2Sn/ KA |
0.98 |
1.1 |
1.0 |
1.1 |
Ph2SnCl2 |
0.35 |
0.31 |
0.30 |
0.33 |
Ph2Sn/ KA |
1.0 |
1.0 |
1.2 |
1.1 |
Cisplatin |
8.3 |
5.4 |
4.3 |
6.6 |
Sample |
EC50 WI-38/
EC50HT-29 |
EC50 WI-38/
EC50MCF-7 |
Me2SnCl2 |
0.39 |
0.39 |
Me2Sn/ KA |
1.0 |
0.91 |
Et2SnCl2 |
0.67 |
0.71 |
Et2Sn/ KA |
0.87 |
0.93 |
Bu2SnCl2 |
0.17 |
0.29 |
Bu2Sn/ KA |
1.00 | 2021 Copyright OAT. All rights reserv
0.96 |
Oc2SnCl2 |
0.46 |
0.43 |
Oc2Sn/ KA |
0.98 |
0.94 |
Ph2SnCl2 |
0.45 |
0.37 |
Ph2Sn/ KA |
0.86 |
0.88 |
Cisplatin |
4.6 |
3.3 |
The CI50 values are similar employing either WI-38 or 3T3 cell as the standard. Thus, it is appropriate for this study to employ either in evaluation of the polymers to inhibit cell growth.
CI50 values greater than one are positive since they indicate a tendency for the test compound to inhibit at concentrations greater than for the standard. For the present study while some show values greater than one none of the polymers show CI50 values much greater than one.
There is not agreement as whether CI50 or EC50 have greater significance in evaluating the ability to inhibit cell growth. For the present study, the polymers show decent EC50 values but no high CI50 values.
Organotin polyesters are formed in low to moderate yield from reaction of the disalt of KA with organotin dichlorides employing the interfacial polycondensation process. All of the reactants are commercially available and the interfacial polycondensation process is employed industrially in the synthesis of polycarbonates and aramid fibers. Thus, a scale-up of the process to produce more of the product is somewhat straight forward. Infrared spectroscopy shows the formation of Sn-O bonds and presence of spectral bonds from both reactants. NMR and MALDI MS are also consistent with the formation of organotin polyesters. MALDI MS shows the presence of 5 to 8 repeat units in the ion fragments with isotopic abundance matches consistent with the presence of tin in the ion fragments. All of the polymers show the ability to inhibit all of the tested human cancer cell lines including two breast and two pancreatic cancer cell lines.
- Carraher C, Roner MR (2014) Organotin polymers as anticancer and antiviral agents. J Organomet Chem 751: 67-82.
- Ott P, Clarsen O, Larsen FS (2005) Cerebral metabolic disturbances in the brain during acute liver failure: from hyperammonemia to energy failure and proteolysis. Neurochemistry International 47:1-2, 13-18.
- Hares P, James IM, Peterson RM (1978) Effect of ornithine alpha ketoglutarate (OAKC) on the response of brain metabolism to hypoxia in the dog. Stroke 9: 222-224.
- Long L, Halliwell B (2011) Artifacts in cell culture: alpha-ketoglutatate can scavenge hydrogen peroxide generated by ascorbate and epigallocatechin gallate in cell culture media. Biochemical and Biophysical Res Comm 406: 20-24.
- Chin RM, Fu X, Pai MY, Vergnes-Heeju L (2014) The metabolite alpha-ketogllutarate extends lifespan by inhibiting ATP synthease and TOR. Nature 510: 397-401.
- Klysz D, Tai X (2015) Glutamine-dependent alpha-ketoglutarate production regulates the balance between T helper 1 cell and regulatory T cell generation. Science signaling 8: ra97.
- Ding P, Huang KL, Li GY, Liu YF (2007) Preparation and properties of modified chitosan as potential matrix materials for drug sustained-release beads. Int J Bioloical Macromolecules 41: 125-131.
- Ding P, Huang KL, Li GY, Zeng WW (2007) Mechanisms and kinetics of chelating reaction between novel chitosan derivatives and Zn(II). J Hazardous Materials 146: 58-64.
- Ma Q, Gao Y, Zou Z (2000) Studies on synthesis and adsorption properties of chitosan modified by ?-ketoglutaric acid. Huaxue Sjoko 22: 327-330.
- Barrett DG, Yousat MN (2008) Poly (triol α-ketoglutarate) as biodegradable, chemoselective, and mechanically tunable elastomers. Macromolecules 41: 6347-6352.
- Barrett DG, Yousat MN (2008) A tunable, chemoselective, and moldable biodegradable polyester for cell scaffolds. ChemBioChem 9: 62-66.
- Deng SJ, Chem J, Qian H, Lin LZ, Zhang N (2015) Synthesis and structure of asymmetrical copper complex and its catalytic performance for styrene epoxidation. Huaue Yanjiu Yu Yinghyong 27: 1549-1553.
- Ferrari MB, Bisceglie F, Fava G, Pelosi G, Giorgio P, et al. (2002) Synthesis, characterization and biological activity of two new polymeric copper(II) complexes with α-ketoglutaric acid shiosemicarbazone. J Inorg Biochemistry 89: 36-44.
- Akbarov AB, Aliev UE, Mutalibov AS (1993) Mixed-ligand complexes of Co+2 and Ni+2 with α-ketoglutaric, aspartic, and glutamic acids. J Inorg Chem 38: 1331-1336.
- Ferri D, Burgi T, Baiker B (2000) Conformational isomerism. A FTIR and ab ignition study. J Chem Soc Perkin Trans 2: 221-228.
- Yolou S, Delarbre JL, Maury L (1992) Spectres de vibration de l’acide α-oxoglutarique et de ses sels alcalins. Can J Applied Spectoscopy 37: 10-17.
- Carraher C, Roner MR, Sooledo N, Moric-Johnson A, Miler L, et al. (2017) Synthesis and Initial Cancer Cell Results of Organotin Polyethers Derived from the Anticoagulant Dicumarol. Int J Applied Pharm Biological Res 2: 1-17.
- Carraher C, Roner MR, Black K, Frank J, Moric-Johnson A, et al. (2017) Polyesters from Reaction of 3,5-Pyridinedicarboxylic Acid and Group V-Containing Dihalides and Their Preliminary and Comparative Ability to Inhibit Cancer Cell Growth. International Journal of Applied Pharmaceutical and Biological Research 2: 1-17.
- Carraher C, Roner MR, Campbell A, Moric-Johnson A, Miller L, et al. (2017) Group IVB Metallocene Polyesters Containing Camphoric Acid and Preliminary Cancer Cell Data. International J Polymeric Materials Polymeric Biomaterials.
- Barber M, Bordoli RS, Sedwick RD, Tyler AN (1981) Fast atom bombardment of solids as an ion source in mass spectroscopy. Nature 293: 270-275.
- Liu LK, Busch KL, Cooks RG (1981) Matrix-assisted secondary ion mass spectra of biological compounds. Analyt Chem 53:109-113.
- Tanaka K, Waki H, Ido Y, Akita S, Yoshida Y, et al. (1988) Protein and Polymer analyses up to m/z 100 000 by Laser Ionization Time-of flight Mass Spectrometry. Rapid Communications in Mass Spectrometry 2: 151–153.
- Karas K, Bachmann D, Hillenkamp F (1985) Influence of the wavelength in high-irradiance ultraviolet laser desorption mass spectrometry of organic molecules. Analytical Chemistry 57: 2935–2939.
- Carraher C, Barot G, Battin A (2009) Reactions between the matrix and ion fragments created from the MALDI MS for organotin-containing polymers. J Polym Mater 26:17-31.
- Carraher C, Sabir T, Carraher CL (2006) Fragmentation matrix assisted laser desorption/ionization mass spectrometry-basics. J Polym Mater 23: 143-151.
- Carraher C, Sabir T, Carraher CL (2008) Fundamentals of fragmentation matrix assisted laser desorption/ionization mass spectrometry. Inorganic Organometallic Macromolecules Springer, NY pp. 329-350.
- Carraher C, Suresh V, Roner MR (2015) Self matrix activity of organotin polyether ester polymers containing alpha-cyano-4-hydroxycinnamic acid. JCAMS 3: 32-44.
- Carraher C, Roner MR, Carraher CL, Crichton R, Black K (2015) Use of Mass Spectrometry in the Characterization of Polymers Emphasizing Metal-Containing Condensation Polymers. J Macromol Sci A 52: 867-886.
- Perera A, Parkes HG, Herz H, Haycock P, Blake DR, et al. (1997) High resolution 1H NMR investigations of the reactivities of alpha-keto acid anions with hydrogen peroxide. Free Radic Res 26: 145-157. [Crossref]
- Sobolev AP, Brosio E, Gianferri R, Segre AL (2005) Metabolic profile of lettuce leaves by high-field NMR spectra. Magn Reson Chem 43: 625-638. [Crossref]






