Tertiary structure and its role in function of a transmembrane protein is still quite difficult to study in modern biology. Here, we propose a new hypothesis that phosphate residues of phospholipids stabilize secondary structure of transmembrane polypeptides by catalyzing a closed-circular resonance reaction of amide bonds across the polypeptide strands on both surfaces of the lipid membrane. The phosphate residue of phospholipid may play a key role in regulation of membrane protein structure, stability, and function.
hydrogen bond, phospholipid, polypeptide, resonance, stabilization, structure
Inside the lipid bilayer, carbonic hydrogens of alkyl groups of fatty acids surround all around the transmembrane polypeptides [1]. Since hydrogen and carbon form a nonpolar covalent bond, alkyl groups interact with polypeptide by hydrophobic interaction [1,2]. Accordingly, hydrophobic side residues locate outside of protein structure to interact with the alkyl groups of fatty acids, while almost all the amide of polypeptides locates inside of the protein structure to form alpha-helix or beta-sheet structures. An alpha-helix contains three hydrogen-bond chains along its long axis [1,2]. An anti-parallel beta sheet forms a barrel-like structure or beta-barrel, which usually contains more than ten hydrogen-bond chains, in a lipid bilayer [1,2]. Hydrogen bonds play a central role to stabilize secondary structure of polypeptide in the lipid bilayer. The hydrogen bonds are stabilized structurally with a resonance effect of amide residues as previously discussed [1-3]. The resonance caused planar structure of amide residues, which not only stabilizes secondary structure of polypeptide thermodynamically but also increases protease-resistant property because disruption of planar structure of amide is required for hydrolytic degradation of a peptide by enzymes [1-3].
Just on the surface of phospholipid bilayer, some amide residues do not participate in hydrogen bonding with anti-parallel strands of a transmembrane beta-barrel and remain free from beta-sheet formation (Figure 1). The free amide residues expose carbonyl oxygen and amino hydrogen to the phosphate residues that locates just upon the hydrophobic layer (Figure 1A). Accordingly, the amino hydrogen could readily make a hydrogen bond with unsaturated oxygen atom of phosphate residues (Figure 1B). On the other hand, although the carbonyl oxygen could make a weak hydrogen bond with hydrogen of phosphate residues, almost all the phosphate groups bind to a monovalent cation such as sodium and potassium in a solution under neutral pH condition (Figure 1B). The phosphate-bound cationic ion is easily released at high frequency to attack any neighboring carbonyl oxygen and induce keto-enol resonance of the amide residues (Figures 1A and 1B). While a hydrogen bond has directional specificity for its strength of association, an ionic interaction could happen at any directions [1,2]. Moreover, phosphate residues interact to each other for the surface conductance of phospholipid bilayer [4]. Taken all together, just upon the surface of hydrophilic alkyl layer phosphate networks surround free amide residues of transmembrane polypeptides and the phosphate group induces keto-enol resonance of the amide residues by releasing its cation to the carbonyl oxygen together with acceptance of amino hydrogen on its unsaturated oxygen (Figure 1B).
It is difficult for an individual amide residue to perform the keto-enol resonance by itself because all the amino hydrogen and carbonyl oxygen form hydrogen bond with the neighboring carbonyl oxygen and amino hydrogen, respectively (Figure 1B). If one amide residue transforms from keto-type to enol-type, all the other amide residues should transform from keto-type to enol-type in a synchronizing manner. How come? Figure 1B shows a possible molecular mechanism which could make a synchronized resonance across the anti-parallel strands of beta-barrel structure. Because keto-enol resonance stabilizes planar structure of amide bonds in polypeptides, the synchronized resonance should stabilize secondary structure of the transmembrane polypeptides.
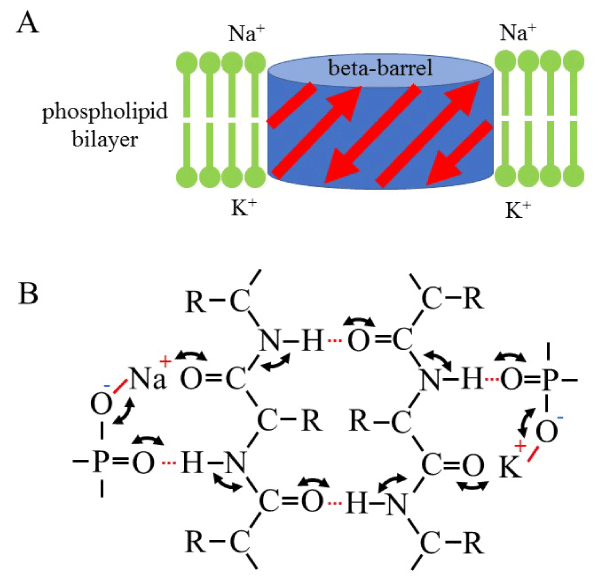
Figure 1. Schematic models of the structure of polypeptides in a phospholipid bilayer. A, Cartoon of beta-barrel structure in a phospholipid bilayer. A red arrow indicates a beta-strand. A light-green line indicates a phospholipid molecule with a hydrophilic head region (circle). B, a closed circular keto-enol resonance reaction among outside and inside phosphate groups of phospholipids and amide bonds of anti-parallel betta-strands of polypeptides. Note that only two of anti-parallel beta-strands were presented here for simplicity. The resonance reactions do not alter the relative position of atoms participated. In this model, left and right side correspond outer- and inside-membrane of a living cell in a depolarizing state, respectively. A black solid line indicates a covalent bond. A red solid line indicates an ionic bond. A red dotted line indicates a hydrogen bond.
In this model, amide residues of transmembrane polypeptides should interact with phosphate residues at both surfaces of phospholipid bilayer. The frequency of synchronized resonance could be regulated by monovalent cations bound to the phosphate groups. Since hydrogen ion stably binds to the phosphate at low pH condition to decrease synchronizing frequency, lower pH may destabilize secondary structure of transmembrane polypeptide. This might be consistent with the fact that some bacterial beta-barrel proteins in the outer-membrane or porins are known to transport small molecules across the membrane in a pH-dependent manner [5].
On the other hand, monovalent cations such as sodium or potassium ions may induce the high-frequency synchronized resonance at neutral condition. Especially, potassium ions may cause higher frequency of resonance than sodium ions judging from the electro-negativity [1,2]. Moreover, if both sides of the phospholipid bilayer are coated with the same species of ions, the resonance of closed circle reaction should be most effective because of its high synchronization. This monovalent-dependent structural stabilization of transmembrane polypeptide might play a role in the depolarization-dependent opening of ion channels such as mitochondrial activity-dependent anion channel [6].
It should be noted that this is just a theoretical hypothesis of a novel feature of phospholipid to stabilize polypeptide second structure in lipid membrane. As mentioned above, hydrophobic side chain of polypeptide also plays a pivotal role in stability of transmembrane peptides. Furthermore, hydrophilic side chains of polypeptide, which are expected to locate inside of the beta-barrel, may also play a central role in the stability. Moreover, some outer loop of polypeptide should play an important role in pH- and depolarization-dependent beta-barrel protein functions. Taken all together, it is necessary to accumulate rigid experimental data from now on for confirmation of the model proposed here in the future.
I would express my special thanks to Managing Editor Sandy Williams for encouragement and introduction to Trends in Research, which aims to open and share various new research idea and data for human benefits and public good.
The author declares no conflict of interest.
- Cox MM, Nelson DL (2012) Principles of Biochemistry, W.H. Freeman; 6th edition.
- Pauling L (1988) General Chemistry, Dover Publications; 3rd edition.
- Aizawa H (2018) On the structure of beta-sheet. J Mol Cell Biol Forecast 1: 1003.
- Aizawa H (2018) Electric biochemistry of sphingomyelin. J Mol Cell Biol Forecast 1: 1001.
- Tamm LK, Hong H, Liang B (2004) Folding and assembly of beta-barrel membrane proteins. Biochim Biophys Acta 1666: 250-263. [Crossref]
- Zeth K (2010) Structure and evolution of mitochondrial outer membrane proteins of beta-barrel topology. Biochim Biophys Acta 1797: 1292-1299. [Crossref]

