Abstract
CYP1A2 belongs to cytochrome P450 super family and mainly expressed in liver. C YP1A2 contributes 11% of total drug metabolism. CYP1A2 enzyme is involved in an NADPH-dependent electron transport chain. CYP1A2 has been shown to metabolize nutrients, antibiotics (ciprofloxacin) and flutamide. Impacts of hormone and gender on the regulation of CYP1A2 have not well documented. Current study reports that CYP1A2 expression is diminished in NF-1 and USF-1 deficient HepG2 cells and further NF-1 and USF-1 regulation is mediated through growth hormone and dexamethasone. GH and dexamethasone controlling the regulation and recruitment of NF-1 and USF-1 on CYP1A2 proximal promoter in a gender-dependent manner in cultured hepatocytes. Additionally, DNA/Protein array identified that NF-1 and USF-1 regulation and nuclear translocation was higher in cultured female adult human hepatocytes and this process was potentiated through a combined action of growth hormone and dexamethasone. Alternatively, ChIP assay demonstrates that the nuclear translocated and CYP1A2 promoter bound NF-1 and USF-1 was abundant in cultured human female hepatocytes in response to GH and dexamethasone regime. The expression of CYP1A2 mRNA and protein correlated with the status of NF-1 and USF-1 in sex-dependent fashion and in response to GH and dexamethasone. Results from the study conclude that CYP1A2 regulation is female dominant and controlled through growth hormone and dexamethasone. This article summarizes that molecular impacts of sex and hormones on regulation of human CYP1A2 . The understanding of sex-dependent regulation of CYPs may help to develop P450 mediated therapies for men and women in clinics.
Key words
dexamethasone, human CYP1A2, growth hormone, nuclear factor-1, upstream stimulatory factor- 1, human hepatocytes
Abbreviations
ChIP, chromatin immunoprecipitation; CYP, cytochrome P450; DMEM, Dulbeccos’s modified Eagle’s medium; NF-1, nuclear factor-1; GH, growth hormone; USF-1, upstream stimulatory factor; AP-1, activator protein 1; AP-2, activator protein 2; PVDF, polyvinylidene diflouride; rhGH, recombinant human growth hormone; Dex; dexamethasone siRNA; small inhibitory RNA.
Introduction
The human genome encodes fifty-seven cytochrome P450s [1]. Cytochrome P450 (CYP) enzymes are the key regulators in the phase I-dependent metabolism of drugs and other xenobiotics, mostly catalyzing oxidations of the substrate, but occasionally also involved in reduction reactions [2,3]. GH secretary patterns are sexually dimorphic and are episodic in men and more continuous in women [4,5]. GH is the major determinant of human CYP regulation [6-8]. Sex-dependent and GH regulated CYPs were very well investigated in rodents models and humans [7,9-19]. Reports strongly suggest that sex dependent and continuous GH induced highest level of CYP3A4 in human primary hepatocytes [7,10]. Similar inherent sexually dimorphic regulation in humans to the same male-dependent GH pattern have been reported for IGF-1, growth and GH binding protein; male>female [20-24]. CYP1A2 is a phase I enzyme and ranks 3rd in total content among all other human hepatic CYPs [25,26].
CYP1A2 is located on human chromosome 15q24.1 and comprising of seven introns and six exons and mainly expressed in the liver, and can be induced via the aryl hydrocarbon receptor (AhR), similarly to CYP1A1 [27,28]. CYP1A2 has been shown to metabolize nutrients, antibiotics (ciprofloxacin) and flutamide [29,30]. Another known substrate for CYP1A2 is caffeine; individuals who carry one or more CYP1A2 *1C alleles are "slow" caffeine metabolizers, whereas carriers of the variant CYP1A2 *1F are "fast" caffeine metabolizers. The same amount of caffeine will therefore tend to have more stimulatory effects on CYP1A2 slow metabolizers than on CYP1A2 fast metabolizer. CYP1A2 has several alleles, which includes CYP1A2 *1, CYP1A2 *1C, CYP1A2 *1D, CYP1A2 *1F, CYP1A2 *1K_-729C>T, CYP1A2 *1K_-739T>G, CYP1A2 *2_63C>G, CYP1A2 *3, CYP1A2 *4, CYP1A2 *6, CYP1A2 *7_3534G>A, CYP1A2 *8, CYP1A2 *11, CYP1A2 *15, CYP1A2 *16 and CYP1A2 _1545T>C. Common polymorphisms with important functional effects in CYP1A2 activity have not been identified and only a couple of very rare genetic variants, CYP1A2 *7 and CYP1A2 *11, have been studied.
GH deficiency leads to reduced activities of CYP1A2 in children and this observation indicates that GH plays a vital role in the regulation of CYP1A2 [30]. Sex dependent regulation of CYP1A2 in humans and pigs has been reported, however the molecular mechanism of sex dependent and GH mediated regulation of CYP1A2 is poorly studied [7,32,33]. Here we cultured primary hepatocytes from men and women under a continuous (feminine) GH regime to elucidate the possible molecular mechanisms associated with the regulation of CYP1A2 . In the present study we show that the regulation of NF-1 and USF-1 transcription factors and their occupancy on CYP1A2 upstream sequence are sexually dimorphic (F>M). Additionally, NF-1 and USF-1 deficient HepG2 cells demonstrate that NF-1 and USF-1 are required for transcriptional activation of the CYP1A2 gene.
Materials and methods
Human Primary Hepatocyte and HepG2 Culture
Hepatocytes were isolated from men and women and plated on rat-tail collagen-coated flasks (T-25) containing Dulbecco’s Modified Eagle’s Medium (DMEM) and were obtained through the Liver Tissue Procurement and Distribution System (Pittsburgh, PA) [34]. All of the samples were obtained with donors consent and with approval of the appropriate hospital ethics committee. Male and female donor varied in age from 21 to 56 years. About 80% were Caucasian; the remaining were African American and Hispanic. Alcohol consumption, smoking and drug history as well as causes of death varied between donors. Approximately 50% of livers had some degree of steatosis (5-40%). Approximately 48 hours after isolation and plating, the primary hepatocyte cultures arrived at our laboratory. HepG2 (ATCC, Manassas, VA) cells were cultured Eagle's Minimum Essential Medium (EMEM). The replacement medium and culture conditions were described previously [7,9,10] .
Hormonal treatments
In order to replicate the more continuous feminine-like growth hormone (GH) profile shown to be favored for human CY1A2 expression, the primary hepatocytes were constantly exposed (2 ng/ml) to rhGH purchased from the National Hormone and Peptide Program (Torrance, CA) [7]. Other cells were exposed to Dexamethasone (Dex) (4 ng/ml) alone or to both Dex and GH. The medium was changed every 12 hours. After 5 days in culture, cells were harvested 60 minutes following the final change of media with hormones as previously reported [7,12]. Cells were exposed to the one of the following conditions, GH or GH diluents or Dex or Dex diluents. (GH-diluent was 0.01 M sodium bicarbonate with and DMSO as dexamethasone diluent.
Preparation of whole cell and nuclear extracts
To isolate total protein for western blots, harvested hepatocytes were centrifuged (800 xg for 10 min) and the resulting cell pellets were re-suspended in lysis buffer [35]. The chromatin was cleared by sonication. Cell lysate was then centrifuged at 12,000 xg for 20 min. The supernatant (whole cell extract) was then removed and stored at -80°C until further analyses. Following, nuclei were isolated as described by a series of centrifugations, re-suspended, homogenized, dialyzed and the nuclear extracts were stored for future analyses. Protein concentrations were determined by Bio-Rad protein assay reagent (Bio-Rad, Hercules, CA) [36].
Bioinformatics analyses
Various bioinformatics tools and programs, including Match-Public (www.gene-regulation.com/cgi-bin/pub/programs/match/match.cgi), AliBaba2 (http://www.generegulation.com/pub/programs/alibaba2/index.html), and Transcription Element Search software, which utilizes the Transcription Factor Database (http://www.cbil.upenn.edu/cgi-bin/tess/tess), were used to identify transcription factor binding sites on the human CYP1A2 upstream sequence.
NF-1 and USF-1 Knockdown in HepG2 Cells
NF-1 and USF-1 knockdown was carried out as described and following the manufacturer’s instructions (Santa Cruz Inc, CA) [37-39]. After 48 hours post transfection of NF-1 siRNA or USF-1 siRNA, the HepG2 cells were treated with 2 ng/ml rhGH (NHPP, Torrance, CA) for 24 hours and medium was replaced every 12 hours [12]. On the 3rd day cells were harvested after 60 minutes following the addition of Dex or GH or combination with Dex and GH. During harvest, HepG2 cells were washed with ice-cold phosphate-buffered saline (PBS). Some of the cells were processed for RNA analysis, ChIP assay and some of the cells were used for nuclear extract preparation. In some experiments, control cells were incubated in the presence of rhGH diluent instead of the hormone or Dex diluent instead of Dex.
RNA analysis
Total RNA was isolated from cultured human hepatocytes or NF-1 or USF-1 proficient or deficient HepG2 cells (NF-1 or US-1 knockdown cells) after hormonal treatments. Five micrograms of total RNA was reverse transcribed with random hexamers. Approximately 25-50 ng of cDNA was applied in real time PCR as described in [40,41]. Real time PCR was performed with an ABI Step-One apparatus using Power SYBR Green Master Mix and using CYP1A2 mRNA forward and reverse primer sequences presented in supplemental table 1. The mRNA signals were normalized with GAPDH mRNA signals (internal control) and quantitated using CT values.
Western blotting analysis
Immunoblotting was performed as described, approximately 25 to 50 µg of total extracts or nuclear extracts from cultured human hepatocytes, NF-1 or USF-1 proficient and deficient HepG2 cells were resolved on 10% SDS-polyacrylamide gel electrophoresis and transferred electrophoretically onto PVDF membranes with a Bio-Rad transfer unit [7,13]. The membranes were then blocked with 5% non-fat dry milk and incubated with primary antibody raised against recombinant human CYP1A2 (kindly provided by Dr. F. Peter Guengerich), β-actin, NF-1 and USF-1 (Santa Cruz, Inc CA). The primary antibody was located by using horseradish peroxidase conjugated to anti-rabbit IgG. The blots, incubated with SuperSignal West Femto (Pierce, Rockford, IL), were visualized, captured and quantified by using an Alpha Innotech FluroChem 8800 Imager (Alpha Innotech, San Leandro, CA) with a movie mode. Signals were normalized to a positive control sample, which was repeatedly run on each blot and exhibited a concentration variant between blots of 3.2 to 6.7% for the different proteins. Lastly, blots were stripped and re-probed with loading control p97 or β-actin antibody and found to be comparable to those obtained with internal controls of the assayed samples.
Protein/DNA array hybridization and analysis
To investigate the relative binding of GH and Dex induced transcription factors to their unique consensus DNA sequences in human hepatocytes, we used TransSignal protein/DNA array (Cat # MA1210, Panomics, Redwood City, CA). In comparison to gel shift assays, which identifies few transcription factors at a time, the TransSignal-Protein/DNA array profiles multiple transcription factor-DNA interactions simultaneously. Array analysis was performed as per the manufacturer's instructions using nuclear extracts from cultured male and female cultured human hepatocytes. Nuclear extracts were prepared using the Panomics nuclear extraction kit (Cat # AY2002) as per the manufacturer's instructions. An equal amount of nuclear extracts (10 µg) from cultured male and female hepatocytes were used for hybridization with TransSignal probe mix (10 µl) containing 56 biotin-labeled double-stranded DNA oligonucleotides. The biotin-labeled DNA binding oligonucleotides were preincubated with the nuclear extracts to allow formation of protein/DNA complexes according to the protocol suggested by the manufacturer of TranSignal Protein/DNA Array System. The protein/DNA complexes were separated from the free probes using a spin column separation system. The bound probes were extracted and hybridized to an array membrane spotted with 56 different consensus-binding sequences following a protocol suggested by the manufacturer. The biotin-labeled oligonucleotides specifically bound to the TFs were extracted and hybridized to the TransSignal array membrane spotted with 56 different consensus-binding sequences for overnight at 42°C. The blots were then washed and incubated with a horseradish peroxidase (HRP)-conjugated streptavidin according to the manufacturer's instructions. The resulting signals were visualized and captured on FluorChem TM IS-8800 Imager by using a movie mode (X-ray film was not used to avoid saturation). The spots were identified with reference provided in the TransSignal Protein/DNA/ Array I kit. The signals were quantitated using FluorChem TM IS-8800 Imager and the signals were normalized with references provided in the TransSignal Protein/DNA Array I kit. The data were quantitatively compared using Sigma Plot.
Chromatin immunoprecipitation assay (ChIP)
Following the hormonal regimen described above, ChIP assays ( 15 ) were performed in primary human hepatocytes as well as HepG2 cells (American Type Culture Collections, HB-8065, Manassa, VA) according to our previously described procedure [13,14]. Nuclei were lysed and the purified nuclei were sonicated to generate DNA fragments with an average length of 200 to 500 bp. Equal concentrations of chromatin from all treatment groups were pre-cleared with protein A agarose beads in the presence of bovine serum albumin and sonicated salmon sperm DNA to reduce the background. After removal of beads by centrifugation, 2 µg of NF-1 or USF-1 specific antibody (Santa Cruz Biotechnology) was added and incubated at 4°C for overnight on a rotary platform. The immunoprecipitates were washed sequentially, eluted and prepared as previously reported [13]. Immunoprecipitated DNA was purified using a PCR purification kit (Qiagen, Valencia, CA) and re-suspended in 50 µl of sterile water. The immunoprecipitated and input DNA were analyzed by semi-quantitative PCR using forward and reverse primers of the CYP1A2 upstream sequence made from the CYP1A2 gene sequence [42-44] (Supplemental Table 1). PCR products resolved on agarose gels were quantified using FluroChem IS-8800 Imager.
Southern blotting analysis
The conventional Southern blotting procedure was applied to confirm the USF1 AND NF-1binding sites with in the ChIP PCR products. Southern blotting was carried out as described before to confirm the NF-1 and USF-1 binding motif in the ChIP PCR products [13,14]. Following, we synthesized the oligonucleotides encompassing the binding sites on the CYP1A2 upstream sequence and labeled this oligonucleotide with γ-32P [42] (Supplemental Table 1). The γ-32P labeled radioactive oligonucleotide served as a probe in Southern blot analysis. Meanwhile, the PCR amplified DNA from the ChIP assay was separated by agarose gels and DNA from the agarose gel was transferred to a nylon membrane; the membrane was baked at 80°C for 2 hours. After 8 hours of pre-hybridization, the membrane was incubated with denatured single-stranded radio labeled probe and hybridized for overnight at 42°C. Membranes were washed with specific buffers and the signals were scanned and quantitated using a FluroChem TM IS-8800 Imager. The signals were normalized to positive controls, which were repeatedly run on each blot
Statistics
All data were subject to analysis of variance. Significant differences were determined with t statistics and the Bonferroni procedure for multiple comparisons.
Results
Mapping of the transcription factor binding sites on the human CYP1A2 upstream sequence
1 Kb CYP1A2 upstream sequence was analyzed using bioinformatics tools to identify the transcription factor binding sites. The bioinformatics tools identified NF-1 and USF-1 (USF-1 close to +1 site and another USF-1 is far from +1 site) putative binding sites along with other transcription factors are presented in the supplemental figure 1. Human CYP1A2 gene consists of seven introns and six exons and exhibits several polymorphic alleles (Supplemental Table 2). NF-1 and USF-1transcription factors that regulate human CYP1A2 [43,44]. Accordingly, we examined the effectiveness of siRNA targeted to NF-1 or USF-1 in HepG2 cells. HepG2 cells transfected with nonspecific (scrambled) siRNA in the presence of Dex and GH combination exhibited the highest level of NF-1 and USF-1 expression and their nuclear translocation. Cells transfected with NF-1 siRNA or USF-1 siRNA showed ~50-60% reduction in NF-1 or USF-1 expression and nuclear translocation and responsiveness to Dex and GH (Figure 1A,B). Similarly, HepG2 cells exposed to Dex alone showed a modest increase in the expression and nuclear translocation of NF-1 or USF-1. NF-1 siRNA or USF-1 siRNA knockdown showed both a ~50-60% lesser expression and nuclear NF-1 or USF-1 under dex condition in the nuclear extracts. HepG2 cells exposed to GH alone showed the smallest increase in expression and nuclear translocation of NF-1 or USF-1, whereas NF-1 siRNA or USF-1 siRNA produced a ~50-60% reduction of expression and nuclear accumulation of NF-1 or USF-1 under GH alone growth condition (Figure 1A,B).In vehicle treated cells, no measurable levels of nuclear NF-1 or USF-1 were observed (data not presented) in nuclear extracts. NF-1 or USF-1siRNA transfected HepG2 reduced the effectiveness of each hormone treatment to stimulate NF-1 and USF-1 expression and nuclear accumulation by ~ 60%. While this level of knockdown is somewhat modest when compared to the effects of other species of siRNA, it is in agreement with reports specifically using siRNA against NF-1 and USF-1 [38,39,45-47]. In general, exposure of the HepG2 cells to the NF-1 or USF-1 siRNA knockdown reduced the effectiveness of each hormone treatment to stimulate NF-1 or USF-1 nuclear translocation by about 60%.
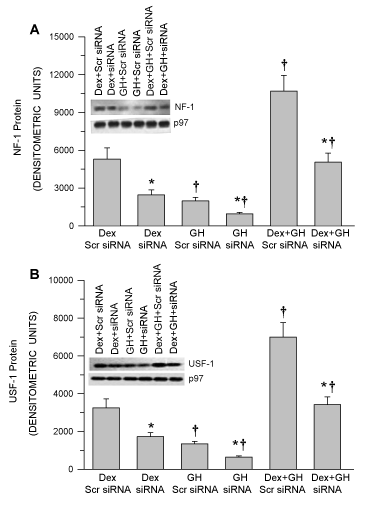
Figure 1. Analysis of hormonal regulation of nuclear NF-1 and USF-1 levels in NF-1 and USF-1 proficient and deficient HepG2 cells. Following 50–70% confluency, HepG2 cells were transfected with either the control non-specific scrambled (Scr) siRNA (proficient) or NF-1 or USF-1 knockdown siRNA (deficient) and 24 hours later the cells were exposed to either continuous dexamethasone alone (Dex) or continuous GH alone (GH) or both hormones (Dex+GH) for 2 days after which the cells were harvested and analyzed. (A) A graphic representation of nuclear NF-1 levels and a representative immunoblot of NF-1 and their respective loading controls. (B) A graphic representation of nuclear USF-1 levels and a representative immunoblot of USF-1 and their respective loading controls. Positive controls for (NF-1 and USF-1) were repeatedly run on all blots for procedural integrity (not shown). Each data point is a mean ± S.D. from 5 or more independent experiments. *P < 0.01 compared the knockdown siRNA with the control cells exposed to the same hormone treatment. †P < 0.01 compared with dexamethasone alone in cells transfected with the same siRNA.
NF-1 or USF-1 deficiency fails to recruit NF-1 or USF-1 on to the CYP1A2 upstream sequence
GH regulated expression levels of NF-1 and USF-1 directly correlated with the nuclear translocated NF-1 or USF-1 and occupancy of these transcription factors on the CYP1A2 upstream sequence. Regardless of the treatment, there was a direct correlation of nuclear levels and the promoter binding levels of NF-1 or USF-1. The greatest binding affinity was observed in Dex and GH combination than other treatment groups (Dex+GH is greater than Dex alone and is greater than GH alone (Figure 2A,3B). NF-1 or USF-1 deficiency leads to weaker recruitment of NF-1 or USF-1 to the CYP1A2 upstream sequence. Further, Southern blotting confirms similar levels of the occupied NF-1 and USF-1 binding motifs of the CYP1A2 upstream sequence as seen in the ChIP assays (Figure 2B,3B). These results clearly demonstrate the effectiveness of NF-1 siRNA and USF-1 siRNA to reduce promoter binding of the nuclear factors by ~ 60%.
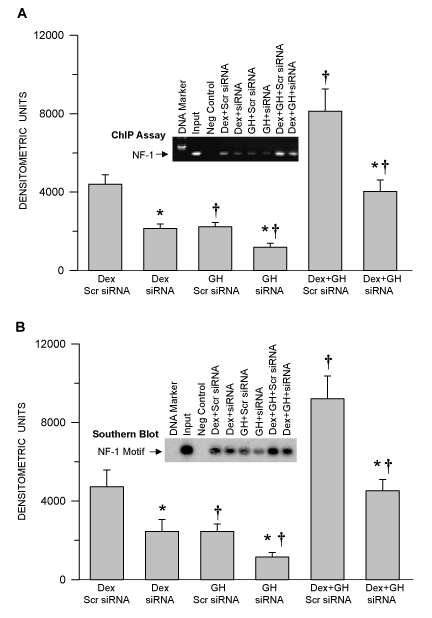
Figure 2. Hormonal regulation of NF-1 binding to the CYP1A2 upstream sequence and confirmation of the occupied NF-1 binding motif in the CYP1A2 upstream sequence in NF-1 proficient and deficient HepG2 cells. Following 50–70% confluency, HepG2 cells were transfected with either control non-specific scrambled (Scr) siRNA (proficient) or NF-1 knockdown siRNA (deficient) and 24 hours later the cells were exposed to either continuous dexamethasone alone (Dex) or continuous growth hormone alone (GH) or both hormones (Dex+GH) for 2 days, after which the cells were fixed with formaldehyde, lysed and the chromatin was sheared and processed for further analysis. (A) A graphic representation of the ChIP-PCR amplified signal and a representative agarose gel picture of the ChIP assay. (B) A graphic representation of the occupied NF-1 binding motif and a representative Southern blot image (insert). Rabbit IgG served as a negative control and input chromatin as a positive control. Each data point is a mean ± S.D. from 5 or more independent experiments. *P < 0.01 compared the effects of the knockdown siRNA with control cells exposed to the same hormone treatment. †P < 0.01 compared with dexamethasone alone in cells transfected with the same siRNA.
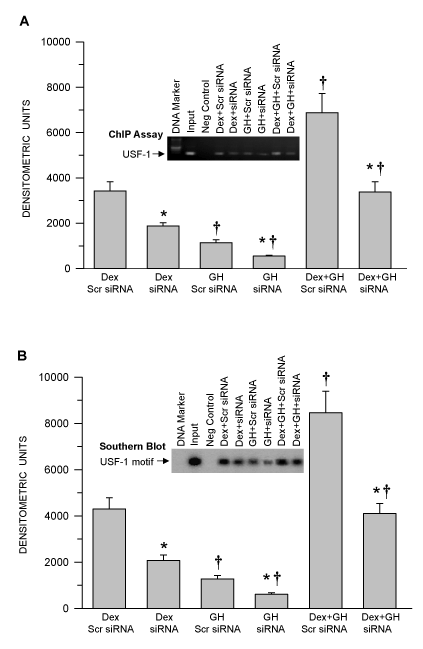
Figure 3. Hormonal regulation of USF-1 binding to the CYP1A2 upstream sequence and confirmation of the occupied USF-1 binding motif in the CYP1A2 upstream sequence in USF-1 proficient and deficient HepG2 cells. Following 50–70% confluency, HepG2 cells were transfected with either control non-specific scrambled (Scr) siRNA (proficient) or USF-1 knockdown siRNA (deficient) and 24 hours. Later the cells were exposed to either continuous dexamethasone alone (Dex) or continuous growth hormone alone (GH) or both hormones (Dex+GH) for 2 days after which the cells were fixed with formaldehyde, lysed and the chromatin was sheared and processed for further analysis. (A) A graphic representation of ChIP-PCR amplified signal and a representative agarose gel picture of the ChIP assay. (B) A graphic representation of the occupied USF-1 binding motif and a representative Southern blot image (insert). Rabbit IgG served as a negative control and the input chromatin as a positive control. Each data point is a mean ± S.D. from 5 or more independent experiments. *P < 0.01 compared the effects of the knockdown siRNA with control cells exposed to the same hormone treatment. †P < 0.01 compared with dexamethasone alone in cells transfected with the same siRNA.
Hormonal regulation of CYP1A2 in NF-1 or USF-1 proficient and deficient HepG2 Cells
Using NF-1 and USF-1 deficient cells, we sought to correlate NF-1 or USF-1 binding on the CYP1A2 upstream sequence with expression levels of CYP1A2 in HepG2 cells under the hormone regimes. GH and dex regulated and nuclear accumulated and CYP1A2 upstream sequence bound NF-1 or USF-1 in NF-1 or USF-1 proficient cells, respectively, directly correlated with expression levels of CYP1A2 mRNA as evidenced by qRT-PCR (Figure 4A,B), Whereas hormone treatments (Dex combination with GH was higher than (more than two fold) Dex alone or GH alone) stimulated nuclear accumulation and promoter bound NF-1 or USF-1 in NF-1 or USF-1 proficient and deficient HepG2 cells (Figure 1A,B), no measurable NF-1 or USF-1 binding to the CYP1A2 upstream sequence was observed in control (diluents) treatment by ChIP assay (data not shown). Nevertheless, interference with NF-1 or USF-1 expression by siRNA knockdown repressed the hormone-induced CYP1A2 mRNA by real time RT-PCR accounts ~60% in HepG2 cells at all hormone regimens (Figure 4A,B). GH alone induced the smallest increase in CYP1A2 mRNA, whereas Dex was more than twice as effective and the combined hormone treatment was clearly the most effective. The signals were normalized to GH alone.
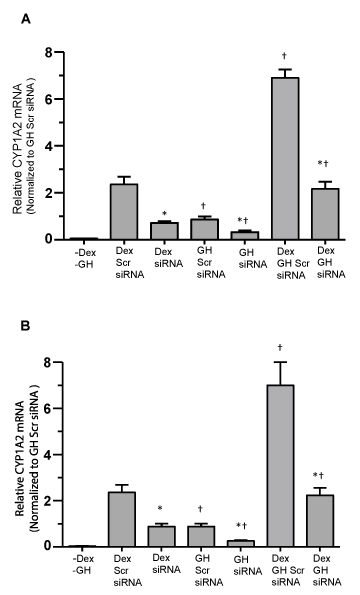
Figure 4. Hormonal regulation of CYP1A2 mRNA levels in NF-1 and USF-1 proficient and deficient HepG2 cells. Following 50–70% confluency, HepG2 cells were transfected with either the control non-specific scrambled (Scr) siRNA (proficient) or NF-1 or USF-1 knockdown siRNA (deficient) and 24 hours later cells were either exposed to continuous dexamethasone alone (Dex) or continuous growth hormone alone (GH) or both hormones (Dex+GH) for 2 days after which the cells were harvested and analyzed. (A) graphic representation of CYP1A2 mRNA levels (qRT-PCR) in NF-1 proficient and deficient HepG2 cells and the signals were normalized with GAPDH mRNA (internal control). (B) A graphic representation of CYP1A2 mRNA levels (qRT-PCR) in USF-1 proficient and deficient HepG2 cells and the signals were normalized with GAPDH mRNA (internal control). Each data point is a mean ± S.D. from 5 or more independent experiments. *P < 0.01 compared the effects of the knockdown siRNA with control cells exposed to the same hormone treatment. †P < 0.01 compared with dexamethasone alone in cells transfected with the same siRNA.
Sex dependent hormonal regulation of CYP1A2 in cultured human hepatocytes
Having demonstrated CYP1A2 regulation is mediated by Dex and GH in HepG2 cells, we wanted to see if the same was true in normal hepatocytes from adult men and women. In agreement with HepG2 cells (Figure 4A,B), hepatic CYP1A2 mRNA by real time qRT-PCR and protein expression was more than two fold when hepatocytes were exposed to the combined treatment of continuous Dex and GH (Figure 5A,B). GH alone was moderately inductive and dexamethasone itself higher than GH alone. Very low CYP1A2 mRNA levels were measured in vehicle treated men and women hepatocytes, but no measurable CYP1A2 protein levels were observed (data not shown). The signals were normalized to male GH alone. Sex differences in the induction of CYP1A2 were indicated by significantly greater than two fold in female hepatocytes than male hepatocytes following all hormonal exposures (Figure 5A,B). The mRNA and protein expressions correlate with each other and exhibit sex-dependent regulation of CYP1A2.
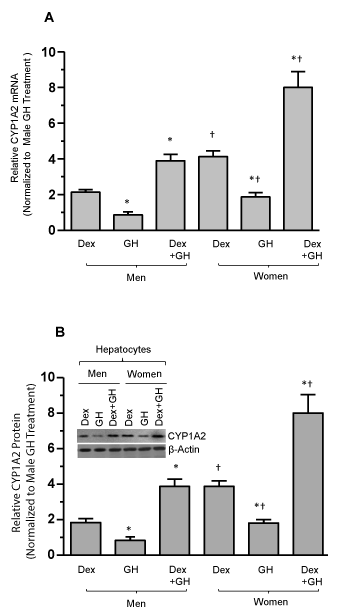
Figure 5. Growth hormone and dexamethasone effects on sex-dependent regulation of human CYP1A2 in cultured primary human hepatocytes. Adult hepatocytes from men and women were either exposed to continuous dexamethasone alone (Dex) or continuous growth hormone alone (GH) or both hormones (Dex+GH) or vehicle (-Dex-GH) alone for 5 days in culture, after which the cells were harvested and processed for further analysis. (A) A graphic representation of relative CYP1A2 mRNA levels in hepatocytes and the signals were normalized with GAPDH mRNA (internal control). (B) A graphic representation of relative CYP1A2 protein levels and a representative immunoblot image of CYP1A2 protein (insert). The protein signal values for vehicle (-Dex-GH) treatments were so low as to be barely detectable and are not included in the figure. Respective loading controls (β-actin) are presented in the figure. Each data point is a mean ± S.D. for cells from 5 or more individuals. *P<0.01 compares to the dexamethasone alone treatment of the same sex. †P<0.01 compares women with men exposed to the same dexamethasone treatment.
Sex dependent regulation of transcription factor levels and binding to nuclear DNA in human hepatocyte culture
In this experiment we investigated whether the hormonally induced transcription factors are sex dependent for their nuclear accumulation and DNA binding activity. We observed that the nuclear concentrations of DNA associated transcription factors AP-1, AP-2, NF-1 and USF-1 were highest levels in female nuclear extracts compared to males exposed to combined GH and Dex (Figure 6A). The Dex treatment alone was modest and the GH was observed as lowest. In all treatment groups the DNA/protein association levels were observed as men < women. Similarly, we observed the nuclear accumulation of NF-1 and USF-1 was greatest in women hepatocytes compared to men exposed to the combined Dex and GH (Figure 6B,C). Induction and nuclear accumulation of NF-1 and USF-1 was modest in Dex alone and least in GH alone. Regardless of treatment, the induction and nuclear accumulation of NF-1 and USF-1 was observed as highest in women hepatocytes. There were no measurable NF-1 and USF-1 in no dexamethasone and no growth hormone growth condition. (-Dex-GH).
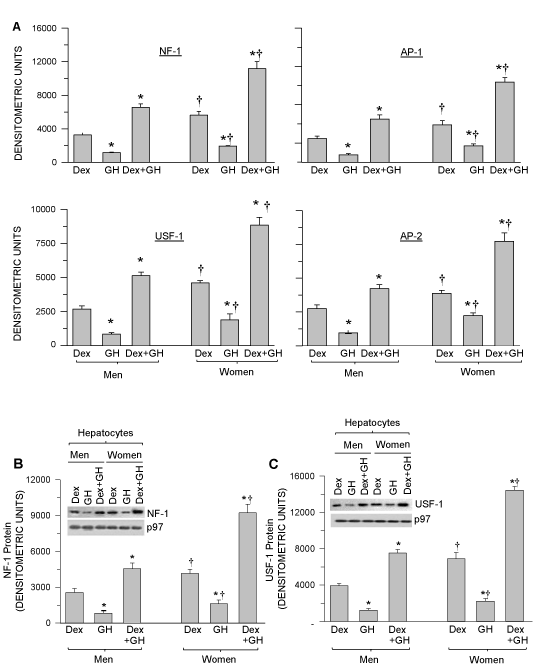
Figure 6. Hormonally regulated, nuclear DNA bound NF-1, USF-1, AP-1 and AP-2 transcription factors in hepatocyte nuclei and nuclear protein levels of NF-1 and USF-1 in cultured hepatocytes from men and women. Adult hepatocytes from men and women were either exposed to continuous dexamethasone alone (Dex) or continuous growth hormone alone (GH) or both hormones (Dex+GH) or vehicle (-Dex-GH) alone for 5 days in culture, after which the cells were harvested and nuclear extract was isolated and processed for further analysis. (A) Graphic representations of nuclear DNA associated transcription factors NF- 1, USF-1, AP-1 and AP-2 levels (Protein DNA array) in the nuclear extracts of cultured male and female hepatocytes (the signals were normalized with the internal controls included in the assay kit). (B) A Graphic representation of nuclear NF-1 protein concentrations and a representative immunoblot image of nuclear NF-1 protein (insert). (C) A Graphic representation of nuclear USF-1 protein and a representative immunoblot image of nuclear USF-1 protein (insert). Respective loading control (p97) for B and C are included in the figure. The protein signal values for vehicle (-Dex-GH) treatment were so low as to be barely detectable and are not included in the figure. Each data point is a mean ± S.D. for cells from 5 or more individuals. *P<0.01 compares to the dexamethasone alone treatment of the same sex. †P<0.01 compares women with men exposed to the same treatment.
Sex-dependent hormonally regulated NF-1 recruitment to the CYP1A2 upstream sequence in men and women hepatocytes
Consistent with the Figure 6A, Dex and GH treatment induced the greatest levels of NF-1 binding to CYP1A2 upstream sequence than either Dex or GH alone (the latter, the least effective) (Figure 7B). NF-1 binding to the CYP1A2 upstream sequence exhibited a sexual dimorphism in all hormone treatments (F>M). In confirmation using Southern blotting, we observed very similar levels of the occupied NF-1 binding motif on the CYP1A2 upstream sequence (Figure 7C) as evidenced by chromatin immunoprecipitation (Figure 7B).
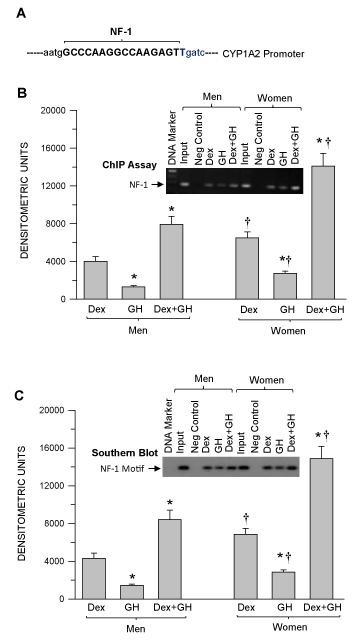
Figure 7. Mapping the NF-1 binding region on human CYP1A2 upstream sequence and sexdependent hormonal regulation of nuclear factor-1 (NF-1) binding to the CYP1A2 upstream sequence as well as confirmation of the occupied NF-1 binding motif in hepatocytes derived from adult men and women. Adult hepatocytes from men and women were either exposed to continuous dexamethasone alone (Dex) or continuous growth hormone alone (GH) or combination of Dex and GH or vehicle (-Dex-GH) alone for 5 days in culture. Next, the cells were fixed with formaldehyde, lysed and the chromatin was sheared and processed for further analysis. (A) NF-1 binding location on the CYP1A2 upstream sequence (Bold). (B) A graphic representation of the ChIP-PCR amplified signal and a representative agarose gel image of the ChIP assay. (C) A graphic representation of the occupied NF-1 binding motif and a representative Southern blot image (insert). Rabbit IgG served as a negative control and input chromatin as a positive control. The ChIP signal values for vehicle (-Dex-GH) treatment were so low as to be barely detectable and are not included in the figure. Each data point is a mean ± S.D. for cells from 5 or more individuals. *P<0.01 compares to the dexamethasone alone treatment of the same sex. †P<0.01 compares women with men exposed to the same treatment.
Sex-dependent hormone regulated USF-1 recruitment to the CYP1A2 upstream sequence in cultured men and women hepatocytes
Consistent with the figure 7B Dex and GH treatments induced the greatest levels of USF-1 binding to the CYP1A2 upstream sequence than either Dex or GH alone (the latter, the least effective) (Figure 8B). USF-1 binding to the CYP1A2 upstream sequence exhibited a sexual dimorphism between all hormone treatments (F>M). In confirmation using Southern blotting, we observed very similar levels of the occupied USF-1 binding motif on the CYP1A2 as evidenced by chromatin immunoprecipitation (Figure 8B).
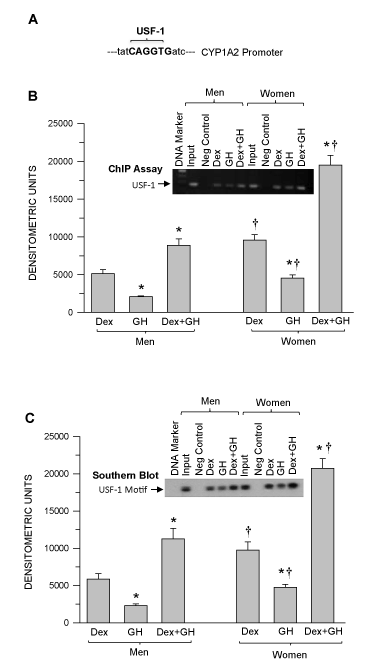
Figure 8. Mapping the USF-1 binding region on the human CYP1A2 upstream sequence and sex-dependent hormonal regulation of USF-1binding to the CYP1A2 upstream sequence and confirmation of the occupied USF-1 binding motif in the CYP1A2 upstream sequence in hepatocytes derived from adult men and women. Adult hepatocytes from men and women were either exposed to continuous dexamethasone alone (Dex) or continuous growth hormone alone (GH) or combination of Dex and GH or vehicle (-Dex-GH) alone for 5 days in culture. Next, the cells were fixed with formaldehyde, lysed and the chromatin was sheared and processed for further analysis. (A) USF-1 binding location on the CYP1A2 upstream sequence (Bold). (B) A graphic representation of the ChIP-PCR amplified signal and a representative agarose gel image of the ChIP assay. (C) A graphic representation of the occupied USF-1 binding motif and a representative Southern blot image (insert). Rabbit IgG served as a negative control and input chromatin as a positive control. The ChIP signal values for vehicle (-Dex-GH) treatment were so low as to be barely detectable and are not included in the figure. Each data point is a mean ± S.D. for cells from 5 or more individuals. *P<0.01 compares to the dexamethasone alone treatment of the same sex. †P<0.01 compares women with men exposed to the same treatment.
Discussion
GH acting through the GH receptor, regulates a complex sexually dimorphic transcriptional program of regulating CYP genes in humans and rodents [7,9-14,48]. The molecular mechanism of GH regulated sex dependent CYP 2A, 2C, and 3A has been well explored [13,49,50]. Human CYP3A4 and CYP3A5 exhibits a female predominance when determined in liver (in vivo) extracts and also in cultured hepatocytes [7,9,10,51-53]. The molecular mechanism of sex dependent regulation of human CYP1A2 is largely unknown and with few reports, which are not describing the sex dependent regulation of CYP1A2 in liver [54,55]. CYP1A2 exhibits in several alleles (Supplemental Table 2). The present study provides a novel molecular mechanism for growth hormone regulated sexually dimorphic expression of CYP1A2. The present study establishes five major key findings; (i) Identification of NF-1 and USF-1 in CYP1A2 gene regulation in response to hormones in HepG2 cells (in vitro). (ii) Activation of NF-1 and USF-1 regulation in response to GH and Dex in adult human hepatocytes. (iii) Identification of sex determined hormonally regulated binding of the NF-1 and USF-1 transcription factors on the CYP1A2 upstream sequence in human hepatocytes. (iv) Confirmation of the NF-1 and USF-1 binding motif on the CYP1A2 upstream sequence. (v) Expression of CYP1A2 transcript and protein expression levels correlates with the relative binding of NF-1 and USF-1 transcription factors in response to sex and hormones.
In the present study, we initially identified the transcription factor binding sites including USF-1 and NF-1 on the CYP1A2 upstream sequence and further we assessed their functional role in the CYP1A2 regulation in response to Dex and GH (Supplemental figure 1). The recruitment of USF-1 and NF-1 to the CYP1A2 upstream sequence are hormonally regulated and their DNA binding are in agreement with earlier findings (Figure 2,3) [42-44]. Interestingly, USF-1 and NF-1 deficiency dictates the repressed expression of CYP1A2 and our data proposes that USF-1 and NF-1 are hormonally regulated and their role is required for CYP1A2 regulation (Figure 4A,B).
In the adult human hepatocyte culture model system, results were in agreement with in HepG2 cells as CYP1A2 regulation was mediated through Dex and GH as well as sex (Figure 5A,B). In agreement with earlier reports, the expression levels of hormonally regulated CYP1A2 are female dominant (F>M) [7,32,33]. These findings support our earlier sex-dependent findings of CYP3A4 and CYP3A5 [9,10]. Nuclear translocation of NF-1 and USF-1 and the association of transcription factors NF-1, USF-1, AP-1 and AP-2 to DNA are Dex and GH regulated and female predominant (Figure 6 A-C). Lastly, the recruitment of NF-1 and USF-1 to the CYP1A2 upstream sequence was female predominant and hormonally regulated (Figure 7 & 8). Both the universal human derived HepG2 model and human primary hepatocyte culture model systems demonstrate that CYP1A2 regulation is mediated through hormonally regulated NF-1 and USF-1 in a sex dependent manner. In addition, several inherent GH-dependent sexually dimorphic responses to non-CYP functions in humans have also been reported (20-23,56). Whereas any possible justification(s) for these intrinsic sexual dimorphisms is speculative, the present study has identified possible mechanisms responsible for CYP1A2 regulation. In summary female-dominant expression of human CYP1A2 appears to be irreversibly inherently expressed. Hormonally regulated and sex dependent activation of transcriptional pathways appear responsible for the sex-dependent expression of CYP1A2. These findings may contribute to the development of sex-dependent therapies.
Acknowledgements
We thank Dr. F. Peter Guengerich (Vanderbilt University School of Medicine) for the generous gift of antibodies against the human isoform CYP1A2 . We also thank Dr. Stephen C. Strom (University of Pittsburgh School of Medicine) for supplying the human hepatocytes through the Liver Tissue Procurement Distribution system funded by the National Institutes of Health Contract N01-DK-9-2310. This work was supported by the National Institute of Health Eunice Kennedy Shriver, National Institute of Health and Human Development [Grant HD-061285].
Author’s contributions
Conception and design: C. Thangavel, E. Boopathi and BH. Shapiro; Development of methodology: C. Thangavel and E. Boopathi; Acquisition of data: C. Thangavel and E. Boopathi; Analysis and interpretation of data: C. Thangavel, E. Boopathi and BH. Shapiro; Computational analysis: C. Thangavel and BH. Shapiro; Writing and review of the manuscript: C. Thangavel and BH. Shapiro.
View supplementary data References
- Guengerich FP (2013) New trends in cytochrome p450 research at the half-century mark. J Biol Chem 288: 17063-17064. [Crossref]
- Guengerich FP (2007) Mechanisms of cytochrome P450 substrate oxidation: MiniReview. J Biochem Mol Toxicol 21: 163-168. [Crossref]
- Guengerich FP (2008) Cytochrome p450 and chemical toxicology. Chem Res Toxicol 21: 70-83. [Crossref]
- Engstrom BE, Karlsson FA, Wide L (1998) Marked gender differences in ambulatory morning growth hormone values in young adults. Clin Chem 44: 1289-1295. [Crossref]
- G. van den Berg, Veldhuis JD, Frolich M, Roelfsema F (1996) An amplitude-specific divergence in the pulsatile mode of growth hormone (GH) secretion underlies the gender difference in mean GH concentrations in men and premenopausal women. J Clin Endocrinol Metab 81: 2460-2467. [Crossref]
- Ahluwalia A, Clodfelter KH, Waxman, DJ (2004) Sexual dimorphism of rat liver gene expression: regulatory role of growth hormone revealed by deoxyribonucleic Acid microarray analysis. Mol Endocrinol 18: 747-760. [Crossref]
- Dhir RN, Dworakowski W, Thangavel C, Shapiro BH (2006) Sexually dimorphic regulation of hepatic isoforms of human cytochrome p450 by growth hormone. J Pharmacol Exp Ther 316: 87-94. [Crossref]
- Liddle C, Goodwin BJ, George J, Tapner M, Farrell GC (1998) Separate and interactive regulation of cytochrome P450 3A4 by triiodothyronine, dexamethasone, and growth hormone in cultured hepatocytes. J Clin Endocrinol Metab 83: 2411-2416. [Crossref]
- Thangavel C, Boopathi E, Shapiro BH (2011) Intrinsic sexually dimorphic expression of the principal human CYP3A4 correlated with suboptimal activation of GH/glucocorticoid-dependent transcriptional pathways in men. Endocrinology 152: 4813-4824. [Crossref]
- Thangavel C, Boopathi E, Shapiro BH (2013) Inherent sex-dependent regulation of human hepatic CYP3A5. Br J Pharmacol 168: 988-1000. [Crossref]
- Thangavel C, Dworakowski W, Shapiro BH (2006) Inducibility of male-specific isoforms of cytochrome p450 by sex-dependent growth hormone profiles in hepatocyte cultures from male but not female rats. Drug Metab Dispos 34: 410-419. [Crossref]
- Thangavel C, Garcia MC, Shapiro BH (2004) Intrinsic sex differences determine expression of growth hormone-regulated female cytochrome P450s. Mol Cell Endocrinol 220: 31-39. [Crossref]
- Thangavel C, Shapiro BH (2007) A molecular basis for the sexually dimorphic response to growth hormone. Endocrinology 148: 2894-2903. [Crossref]
- Thangavel C, Shapiro BH (2008) Inherent sexually dimorphic expression of hepatic CYP2C12 correlated with repressed activation of growth hormone-regulated signal transduction in male rats. Drug Metab Dispos 36: 1884-1895. [Crossref]
- Aparicio O, Geisberg JV, Sekinger E, Yang A, Moqtaderi Z, et al. (2005) Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr Protoc Mol Biol Chapter 21 21: 23. [Crossref]
- Shapiro BH, Agrawal AK, Pampori NA (1995) Gender differences in drug metabolism regulated by growth hormone. Int J Biochem Cell Biol 27: 9-20. [Crossref]
- Pampori NA, Shapiro BH (1999) Gender differences in the responsiveness of the sex-dependent isoforms of hepatic P450 to the feminine plasma growth hormone profile. Endocrinology 140, 1245-1254. [Crossref]
- Waxman DJ, Pampori NA, Ram PA, Agrawal AK, Shapiro BH (1991) Interpulse interval in circulating growth hormone patterns regulates sexually dimorphic expression of hepatic cytochrome P450. Proc Natl Acad Sci U S A 88: 6868-6872. [Crossref]
- Agrawal AK, Shapiro BH (2000) Differential expression of gender-dependent hepatic isoforms of cytochrome P-450 by pulse signals in the circulating masculine episodic growth hormone profile of the rat. J Pharmacol Exp Ther 292, 228-237. [Crossref]
- Soares DV, Conceicao FL, Brasil RR, Spina LD, Lobo PM, et al. (2004) Insulin-like growth factor I levels during growth hormone (GH) replacement in GH-deficient adults: a gender difference. Growth Horm IGF Res 14: 436-441. [Crossref]
- Span JP, Pieters GF, Sweep FG, Hermus AR, Smals AG (2001) Gender differences in rhGH-induced changes in body composition in GH-deficient adults. J Clin Endocrinol Metab 86: 4161-4165. [Crossref]
- Kuromaru R, Kohno H, Ueyama N, Hassan HM, Honda S, et al. (1998) Long-term prospective study of body composition and lipid profiles during and after growth hormone (GH) treatment in children with GH deficiency: gender-specific metabolic effects. J Clin Endocrinol Metab 83, 3890-3896. [Crossref]
- Burman P, Johansson AG, Siegbahn A, Vessby B, Karlsson FA (1997) Growth hormone (GH)-deficient men are more responsive to GH replacement therapy than women. J Clin Endocrinol Metab 82: 550-555. [Crossref]
- Johansson AG (1999) Gender difference in growth hormone response in adults. J Endocrinol Invest 22, 58-60. [Crossref]
- Rodriguez-Antona C, Ingelman-Sundberg M (2006) Cytochrome P450 pharmacogenetics and cancer. Oncogene 25: 1679-1691. [Crossref]
- Guengerich FP (2003) Cytochromes P450, drugs, and diseases. Mol Interv 3: 194-204. [Crossref]
- Ikeya K, Jaiswal AK, Owens RA, Jones JE, Nebert DW, et al. (1989) Human CYP1A2: sequence, gene structure, comparison with the mouse and rat orthologous gene, and differences in liver 1A2 mRNA expression. Mol Endocrinol 3, 1399-1408. [Crossref]
- Fukunaga BN, Probst MR, Reisz-Porszasz S, Hankinson O (1995) Identification of functional domains of the aryl hydrocarbon receptor. J Biol Chem 270: 29270-29278. [Crossref]
- Shet MS, McPhaul M, Fisher CW, Stallings NR, Estabrook RW (1997) Metabolism of the antiandrogenic drug (Flutamide) by human CYP1A2. Drug Metab Dispos 25: 1298-1303. [Crossref]
- Drusano GL, Standiford HC, Plaisance K, Forrest A, Leslie J, et al. (1986) Absolute oral bioavailability of ciprofloxacin. Antimicrob Agents Chemother 30: 444-446. [Crossref]
- Kennedy MJ, Davis DA, Smith N, Gaedigk A, Pearce RE, et al. (2008) Reduced activities of cytochrome P450 1A2 and xanthine oxidase in children with growth hormone deficiency. Clin pharmacology Ther 84: 674-678. [Crossref]
- Kojima M, Sekimoto M0, Degawa M (2010) Androgen-mediated down-regulation of CYP1A subfamily genes in the pig liver. J Endocrinol 207: 203-211. [Crossref]
- Kojima M, Sekimoto M, Degawa M (2008) A novel gender-related difference in the constitutive expression of hepatic cytochrome P4501A subfamily enzymes in Meishan pigs. Biochem Pharmacol 75: 1076-1082. [Crossref]
- Strom SC, Pisarov LA, Dorko K, Thompson MT, Schuetz JD, et al. (1996) Use of human hepatocytes to study P450 gene induction. Methods Enzymol 272: 388-401. [Crossref]
- Garcia MC, Thangavel C, Shapiro BH (2001) Epidermal growth factor regulation of female-dependent CYP2A1 and CYP2C12 in primary rat hepatocyte culture. Drug Metab Dispos 29: 111-120. [Crossref]
- Dignam JD, Lebovitz RM, Roeder RG (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475-1489. [Crossref]
- Chen L, Shen YH, Wang X, Wang J, Gan Y, et al. (2006) Human prolyl-4-hydroxylase alpha(I) transcription is mediated by upstream stimulatory factors. J Biol Chem 281, 10849-10855.
- Gaussin A, Modlich U, Bauche C, Niederländer NJ, Schambach A, et al. (2012) CTF/NF1 transcription factors act as potent genetic insulators for integrating gene transfer vectors. Gene Ther 19: 15-24. [Crossref]
- Waki H, Nakamura M, Yamauchi T, Wakabayashi K, Yu J, et al. (2011) Global mapping of cell type-specific open chromatin by FAIRE-seq reveals the regulatory role of the NFI family in adipocyte differentiation. PLoS Genet 7: e1002311. [Crossref]
- Thangavel C, Boopathi E, Ciment S, Liu Y, Neill OR, et al. (2014) The retinoblastoma tumor suppressor modulates DNA repair and radioresponsiveness. Clin Cancer Res 20: 5468-5482. [Crossref]
- Brandin H, Viitanen E, Myrberg O, Arvidsson AK (2007) Effects of herbal medicinal products and food supplements on induction of CYP1A2, CYP3A4 and MDR1 in the human colon carcinoma cell line LS180. Phytother Res 21: 239-244. [Crossref]
- Quattrochi LC, Tukey RH (1989) The human cytochrome Cyp1A2 gene contains regulatory elements responsive to 3-methylcholanthrene. Mol Pharmacol 36: 66-71. [Crossref]
- Pickwell GV, Shih H, Quattrochi LC (2003) Interaction of upstream stimulatory factor proteins with an E-box located within the human CYP1A2 5'-flanking gene contributes to basal transcriptional gene activation. Biochem Pharmacol 65, 1087-1096. [Crossref]
- Narvaez MJ, Anderson GR, Pickwell GV, Quattrochi LC (2005) Characterization of adjacent E-box and nuclear factor 1-like DNA binding sequence in the human CYP1A2 promoter. J Biochem Mol Toxicol 19: 78-86. [Crossref]
- Ravichandran V, Major EO (2008) DNA-binding transcription factor NF-1A negatively regulates JC virus multiplication. J Gen Virol 89: 1396-1401. [Crossref]
- Wang R, Liang H, Li H, Dou H, Zhang M, et al. (2014) USF-1 inhibition protects against oxygen-and-glucose-deprivation-induced apoptosis via the downregulation of miR-132 in HepG2 cells. Biochem Biophys Res Commun 446, 1053-1059. [Crossref]
- Dickson ME, Tian X, Liu X, Davis DR, Sigmund CD (2008) Upstream stimulatory factor is required for human angiotensinogen expression and differential regulation by the A-20C polymorphism. Circ Res 103, 940-947. [Crossref]
- Sugathan A, Waxman DJ (2013) Genome-wide analysis of chromatin States reveals distinct mechanisms of sex-dependent gene regulation in male and female mouse liver. Mol Cell Biol 33: 3594-3610. [Crossref]
- Waxman DJ, O'Connor C (2006) Growth hormone regulation of sex-dependent liver gene expression. Mol Endocrinol 20: 2613-2629. [Crossref]
- Shapiro BH, MacLeod JN, Pampori NA, Morrissey JJ, Lapenson DP, et al. (1989) Signalling elements in the ultradian rhythm of circulating growth hormone regulating expression of sex-dependent forms of hepatic cytochrome P450. Endocrinology 125: 2935-2944. [Crossref]
- Gleiter CH, Gundert-Remy U (1996) Gender differences in pharmacokinetics. Eur J Drug Metab Pharmacokinet 21: 123-128. [Crossref]
- Jaffe CA, Turgeon DK, Lown K, Demott-Friberg R, Watkins PB (2002) Growth hormone secretion pattern is an independent regulator of growth hormone actions in humans. Am J Physiol Endocrinol Metab. 283: E1008-1015. [Crossref]
- Wolbold R, Klein K, Burk O, Nussler AK, Neuhaus P, et al. (2003) Sex is a major determinant of CYP3A4 expression in human liver. Hepatology 38, 978-988. [Crossref]
- Relling MV, Lin JS, Ayers GD, Evans WE (1992) Racial and gender differences in N-acetyltransferase, xanthine oxidase, and CYP1A2 activities. Clin Pharmacol Ther. 52: 643-658. [Crossref]
- Rasmussen BB, Brosen K (1996) Determination of urinary metabolites of caffeine for the assessment of cytochrome P4501A2, xanthine oxidase, and N-acetyltransferase activity in humans. Ther Drug Monit. 8: 254-262. [Crossref]
- Johansson AG, Engstrom BE, Ljunghall S, Karlsson FA, Burman P (1999) Gender differences in the effects of long term growth hormone (GH) treatment on bone in adults with GH deficiency. J Clin Endocrinol Metab 84: 2002-2007. [Crossref]








