|
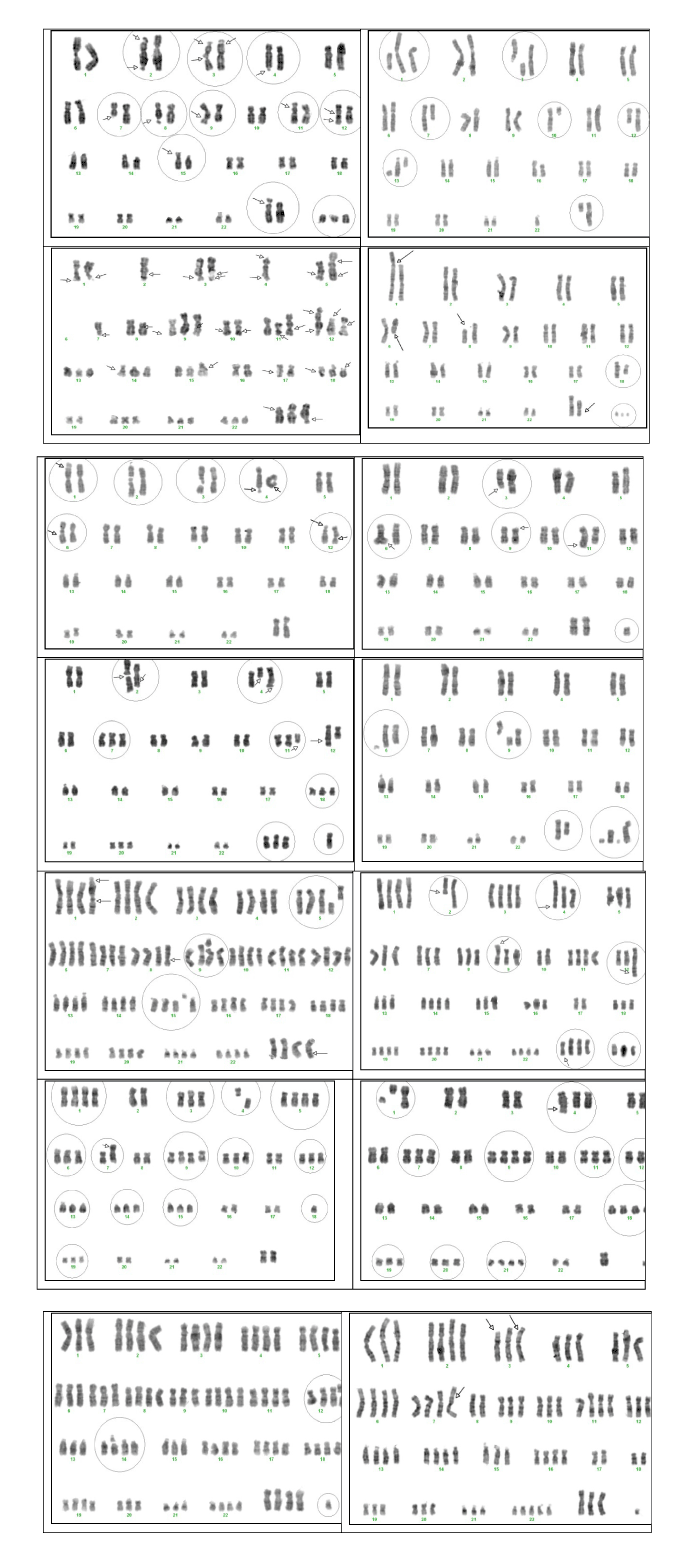
Cells |
Karyotypes |
| |
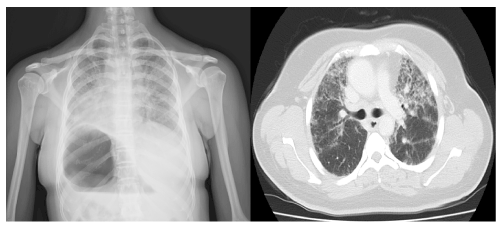
The patient (Girl) |
|
1 |
46,XX,fra(4q33) |
|
2 |
46,XX,del(6q13-qter),del(Xq28-qter) |
|
3 |
93,XXXX,der(10)t(4;10)(q13;p24) |
|
4 |
45,XX,-13,del(18q22-qter) |
|
5 |
47,XX,+mar, dic(4) |
|
6 |
46,XX,del(4q21-qter) |
|
7 |
42,XX,-1(x2),-2,-20,t(1;4)(q12;q34), t(5;20)(q31;p13),del(9q22,2-qter) |
|
8 |
64,XX,+3,+4,+5,+6,+8,+9,+10,+11,+12,+13,+14,+15,+16,+19(x2),+21(x2),+22(x2),t(17;18)(q21;p11,3) |
|
9 |
59,XX,+1(x2),+2,-4,+5(x2),+6(x2),-7,+9,+11, +12,+13,+14, +15,-18,+19, +20,+ace, del(4q25-qter) |
|
10 |
46,XX,chtbr(11p11) |
|
11 |
48,XX,+ace(x2), del(16q23-qter) |
|
12 |
93,XXXX,+20,del(9q13-qter) |
|
13 |
46,XX,fra(8q22),fra(18q23) |
|
14 |
46,XX,fra(6q21,3) |
|
15 |
64,XXX,+1,+4,+6(x2),+7(x2),+8,+9,+10(x2),+12(x2),+17,+18,+19,+20(x2),+X, fra(4q31,3), fra(11q23,2) |
|
16 |
47,XX,+21,dup(7q), del(Xq25-qter) |
|
17 |
91,XXXX,t(8,14)(q24,3;q11,2) |
|
18 |
46,XX,4p+ |
|
19 |
70,XXX,+1,-2,+3(x3),+4,+5(x2),+7(x2),+9(x2),10,+12,+13(x2),+14,+15,+17,+18,+20,+21,+22(x3),+X, der(3)(3;?)(q21;?) |
|
20 |
45,XX,+9,-20,der(4)(4;6)(q31;q13),del(4q32-qter) |
|
21 |
93,XXXX,+Y(?) |
|
22 |
49,XX,+mar,+Y(?),der(3)(3;?)(q27;?),fra(17q21) |
|
23 |
45,XX,-2,chtbr(1p22),chtbr(3q22),del(7q22-qter),der(11),del(12q21-qter),chtbr(13q22),del(Xq12q-ter), del(10q11-qter) |
|
24 |
87,XXXX,+1,+2(x2),+3(x2),+4(x2),+5(x2),+6,+7,+8(x2),+9,+10(x2),+11(x2),12(x2),+13,+14(x2),+15,+16(x2),+17(x2),+18(x2),+19(x2),+20,+21,+22(x2),+X(x2),+mar, chrbr(12q12), chrbr(14q12) |
|
25 |
76,XXX,+1,+2(x2),+3,+4,+5,+6(x2),+7(x2),+9,+10,+11(x2),+12,+13(x2),+14(x2),+15,+16(x2),+19,+20,+21,+22(x3),+X,+mar, t(3;7)(p21,2;p22), fra(3p25) |
|
26 |
49,XX,+18,+ace(x3),t(1;X)(p36,2;p11,2), t(8;X)(p11,2;p13),t(18;18)(q22;q13.1), del(6q16-qter) |
|
27 |
92,XXXX,fra(1p36,1),fra(1q25), chrbr(5q15), fra(8q22),chrbr(9p13),chrbr(15q11), fra(Xq22) |
|
28 |
46,XX,fra(1p22,3),fra(18q21),del(6q1--qter), fra(10p13) |
|
29 |
52,XX,+?,+ace(x3),chrbr(6q25),chrbr(9q22), del(Xq22-qter) |
|
30 |
46,XX,t(2;16)(q31;p13,3),fra(4q33) |
|
31 |
47,XX,+mar,del(3q22-qter), t(3;11)(q22-ter;q21), t(6;11)(q22;q21), del(9p-),qr(6), |
|
32 |
46,XX,t(22;?)(p11;?) |
|
33 |
46,XX |
|
34 |
59,X,+4,+7,+9(x2),+11,+12,+18(x2),+19,+20,+21(x2),+mar(x3),t(1;4)(q23;q35) |
|
35 |
92,XXXX, chrbr(3q24,2), fra(Xq26), 12q+ |
|
36 |
46,XX,fra(2q13)(x2),chtbr(3q27),fra(7q22), fra(Xq23) |
|
37 |
46,XX,fra(1q36,1),chrb(2p22),chrbr(3q21), fra(4q33)(x2),fra(6q13), fra(12p13), chrbr(12q22) |
|
38 |
87,XXXX,-2,-9,-18,-19,-1,fra(10p13), fra(Xq26) |
|
39 |
46,XX |
|
40 |
49,XX,+18,+mar(x2),fra(Xq27),fra(17q21) |
|
41 |
44,XX,-8,-18,-19 |
|
42 |
46,XX,fra(4q33),fra(6q23) |
|
43 |
49,XX,+21,+ace(x2),chrbr(8p23),der(12)t(12;?)(p15;?) |
|
44 |
46,XX |
|
45 |
44,XX,-8,-19,der(5)(p?) |
|
46 |
47,XX,+ace,del(3p22-pter) |
|
47 |
40,XX,-3,-6,-10,-14,-17,-22,del(2q33-qter) |
|
48 |
46, XX,der(9) dup(9p) |
|
49 |
47,XX,-10,+(?)(x2), fra(4q22), fra(4q35), del(11p1-pter),del(11q14,2-qter), del(Xq23-ter) |
|
50 |
86, XXXX,+1,+2(x2),-,+4(x2),+5(x2),6(x2),+7,+8(x2),+9,+10(x2),+11(x2),+12,+13(x2),+14(x2), +15(x3), +16(x2),+17,+18(x2),+19(x2),+20,+21,+22(x2),+X(x2),+mar, der(3)(3q22-qter?), fra(4q35), fra(Xp21) |
|
51 |
77,XXXX,+1(x2),+3(x2),+4,+5,+6,+7,+8,+9,+11(x2),+12,+13,+14(x2),+15,+16,+18,+19(x2), +20(x2),+21, +22(x2), +X(x2),+?(x3), del(2q23),der(4)t(4;?)(q31;?), der(9)t(p24;?), der(12)t(12;7)(24,q;p13), del(Xq22) |
|
52 |
44,XX,-8,-13,der(1)(p36,1;8?), chrbr(3p21.1) |
|
53 |
49,XX,+ace(x3),fra(2p23),fra(2q25),fra(3p25),chrb(3q13,2),fra(3p25),fra(4q25),del(7q22),fra(8q24,1), der(9)t(9;?)(q32;?),der(11)t(11;?)(p11,2;?),fra(12q15),fra(12q24,1), 15p+, chrb(Xp21,2) |
| |
Mother |
| |
46,XX,chrb(11p15); 46,XX,fra(1q42); 48,XX,+11,+?,der(17)t(17;?)(q25;?),fra(5q25); |
| |
44,XX,-10,-14; 46,XX,del(14q32.1),fra(1p32); 41,XX,der(2)t(2;18)(q35;p11.3),der(2)t(2;9); |
| |
45,XX,-21; 46,XX,fra(Xq27); 46,XX,fra(2q21),fra(2q31); 44,XX,-5,-20,der(5)t(5q;?); 46,XX,fra(2q31); 45,XX,-16; 46,XX,fra(11p15); 46,X,-X,+mar; 41,X,-X,-8,-11,-17,-19; 47,XXX; 47,XXX; 47,XXX; 46,XXX,-13,fra(2q31); 46,Xi(Xq) |
| |
Father |
| |
45,XY,-22; fra(1q23); fra(1q32); fra(Xq26); fra(5q23); fra(7q21); gap(13q13) |