Abstract
HPV16 DNA associated with histologic high-grade precancer lesions (cervical intraepithelial neoplasia grade 2 or 3 [CIN2 or CIN3]) is sometimes found in women with atypical squamous cells of undetermined significance (ASCUS) Pap smears, the mildest form of cellular abnormality in a cervical smear. This study evaluated, in a population of patients with ASCUS Pap smears, the prevalence of different HPV16 lineages (or isolates) and the physical status, total viral load, and integrated viral load of HPV16 DNA in a population, stratified by HPV16 isolates (primarily either EUR-350T or EUR-350G) that was found. We demonstrated, in smears of women diagnosed with ASCUS and infected with EUR-350G HPV16 isolates (sublineage A1) that both the physical status of HPV16 and the distribution of integrated HPV16 DNA viral loads were different when compared to a population infected with EUR-350T HPV16 isolates. We showed that the mean and median percentages of HPV16 DNA were higher, and a distribution of log10 integrated HPV16 viral load was more homogeneous in a population infected with EUR-350G HPV16 isolates than in a population infected with EUR-350T HPV16 isolates. No difference was found in terms of total HPV16 viral load between these two isolates. Here, we investigate E6-E7 sequencing as an easy technique to detect women with a greater proportion of integrated HPV genome, and we suggest an adaptation of the current protocol for monitoring women with ASCUS Pap smears.
Key words
HPV, ASCUS, lineage, variant, integration
Introduction
Mucosal Human Papillomaviruses (HPVs) play a crucial role in the development of malignancies. These viruses are responsible for 99.7% of cervical cancers [1] and are classified as High-Risk (HR) or Low-Risk (LR), based on their association to cervical cancer [2,3]. HPV infection is extremely frequent, more often transient, asymptomatic, and cleared in 9-15 months; only a small fraction of women infected with a HR-HPV develop precancerous lesions that progress to cervical carcinoma. Persistent HR-HPV infection (especially HPV16) has been well established as the central cause of cervical cancer and integration of HPV DNA into the host cell genome is believed to be essential for malignant transformation. However, persistent infections with certain variants of HPV16, the genotype found in approximately half of all cervical cancers [4], differ in cancer risk. These variants diverge in their biological properties; therefore, they may become important risk factors in cervical cancer because of possible differences in pathogenicity [5-9].
By convention, the establishment of a new viral type occurs if the nucleotide sequence of the gene encoding the L1 major capsid protein is at least 10% different from all other classified HPV types [2,10]. HPV variant lineages have been found to differ by 1%–10% from full genomes of the same HPV type, and from variant sublineages by 0.5%-1.0% [11,12]. HPV16 variants, representing further evolutionary divergence and each with a different geographic distribution, are classified into four major lineages: A (previously called European-Asian, EAS), B (African 1, Afr1), C (African 2, Afr2), and D (North-American/Asian-American, NA/AA). Furthermore, three European sublineages [A1, A2, A3 (European, E)] and six non-European sublineages [A4 (Asian, As), B1 (Afr1a), B2 (Afr1b), D1 (NA), D2 (AA2), and D3 (AA1)] have been described [12,13]. The sequence analysis of sublineage A1 sometimes shows one nucleotide modification that is localized in the E6 oncogene, leading to an amino acid substitution from leucine to valine (L83V). Thus, sublineage A1 can be divided into isolates containing either 350T (EUR-350T, which includes the prototype HPV16 sequence) or 350G (EUR-350G). Epidemiological studies have suggested that this polymorphism might promote persistent viral infection and cervical cancer development [8,9,14,15]: HPV16 variants with mutations in E6 have been reported for cervical High-grade Squamous Intraepithelial Lesions (HSIL), invasive cervical cancer, and anal HSIL [16-20]. More precisely, in epidemiological study of the French population, a specific intratype polymorphism, such as the EUR-350G isolate, has been found to induce a greater risk of persistence and progression than the E6 prototype [8]. Furthermore, detection of integrated HPV and HPV16 viral loads might also constitute promising markers for cervical disease undergoing rapid progression [21-24].
HPV DNA integration into the host cell genome is believed to be essential for malignant transformation [25-27]. Correlations between the progress of cancer and the physical states of the HPV genome have been shown in different studies; episomal HPV seems to be predominant in the early stages of CIN, while the integrated virus is more frequently detected in high-grade CIN and Squamous Cell Carcinoma (SCC) [27,28]. Nevertheless, a study by Peitsaro et al. also supports the idea that integrated HPV could likewise be found in low-grade lesions (< CIN2) [22].
Another marker of HPV16 infection, carcinogenic high viral load, has been associated with a risk of developing CIN2 or 3 lesions [29-32], whereas other reports showed no correlation [33,34]. Carcopino et al. demonstrated, in cervical smears collected before colposcopic examinations and in women aged 30-40, significant correlations between high HPV16 viral loads and the presence of high-grade squamous intraepithelial lesions [35]. Another study, with a prospective cohort design, reported higher HPV16 viral loads associated with modest increases in the incidence of cytological abnormalities, and predictive of incident CIN2/3+ [36].
The goal of our study was to describe the prevalence of different HPV16 lineages and isolates in women with Atypical Squamous Cells of Undetermined Significance (ASCUS) Pap smears living in the southeast of France. We also aimed to quantify HPV16 physical status and HPV16 DNA viral loads in this population, stratified by HPV16 isolates found. Lastly, based on this study, we may propose an adaptation of the current protocols for monitoring the patient population undergoing ASCUS Pap smears.
Results
During 2012-2015, 106 women with HPV16-positive ASCUS Pap smears attended the University Hospital of Nice (62 women mono-infected HPV16, and 44 infected with HPV16 and one other HR-HPV). Their ages ranged 19-69 years (average 35.3 years).
E6-E7 nucleotide sequences analysis
After a first step of cytological analysis and HPV genotyping, samples diagnosed both as ASCUS (by the Pathology Laboratory of the University Hospital of Nice) and HPV16-positive with PapilloCheck® HPV DNA chips (by the Laboratory of Virology of the University Hospital of Nice) were further amplified to sequence the region containing the E6 and E7 genes (nt 83-858). HPV16 DNA nucleotide positions were numbered according to the published sequence of the reference clone [37]. The alignment of E6/E7 nucleotide sequences (nt 83-858) were referred to the prototype HPV16 sequence (NCBI Reference Sequence: NC_001526.2) using the BioEdit Sequence Alignment Editor. Sequence differences between E6-E7 genes of these HPV16 genotypes are summarized in table 1.
Table 1. Sequence alterations, relative to the E6-E7 ORF of the reference HPV16 sequence (referred to as the prototype HPV16 sequence): each letter represents the nucleotide change at that position. Lower case letter = nucleotide change at that position without an amino acid change. Upper case letter = nucleotide change at that position with a resulting amino acid change. In the “amino acid” column, the letter preceding the amino acid position refers to the reference HPV16 sequence, and the letter after it refers to the substitution. Bold letters indicate an EUR-350G isolate with other amino acid changes.
|
E6 |
E7 |
|
|
|
|
|
|
|
1 |
1 |
1 |
1 |
2 |
2 |
2 |
2 |
2 |
3 |
3 |
3 |
4 |
4 |
4 |
5 |
|
5 |
6 |
6 |
7 |
7 |
7 |
8 |
|
|
|
|
|
|
|
|
0 |
3 |
4 |
4 |
3 |
5 |
6 |
8 |
8 |
3 |
5 |
6 |
0 |
0 |
4 |
3 |
|
9 |
4 |
8 |
3 |
8 |
9 |
2 |
|
|
|
|
|
|
|
|
9 |
2 |
3 |
5 |
9 |
7 |
1 |
6 |
9 |
5 |
0 |
8 |
3 |
6 |
8 |
2 |
|
4 |
7 |
9 |
2 |
9 |
5 |
2 |
|
|
|
|
|
|
|
NC_001526.2 |
T |
G |
C |
G |
G |
A |
T |
T |
A |
C |
T |
G |
A |
T |
G |
A |
|
T |
A |
G |
T |
T |
T |
A |
|
|
|
|
|
|
|
Lineage/isolate |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Amino
Acid |
|
|
|
|
|
|
|
Amino
Acid |
Nb |
% of
variants |
Mean
age |
% of
integration |
Viral
load |
Integrated
viral load |
A-350T |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
- |
|
|
|
|
|
|
|
- |
35 |
37.7 |
35.7 |
25 |
35 c/c |
7 c/c |
A-109c/350T |
c |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
- |
|
|
|
|
|
|
|
- |
1 |
A-350T/822g |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
- |
|
|
|
|
|
|
g |
- |
4 |
A-350G |
|
|
|
|
|
|
|
|
|
|
G |
|
|
|
|
|
L83V |
|
|
|
|
|
|
|
- |
52 |
57.55 |
35.1 |
36.3 |
16 c/c |
6 c/c |
A-350G/594c |
|
|
|
|
|
|
|
|
|
|
G |
|
|
|
|
|
L83V |
c |
|
|
|
|
|
|
- |
1 |
A-350G/689R |
|
|
|
|
|
|
|
|
|
|
G |
|
|
|
|
|
L83V |
|
|
R |
|
|
|
|
-/G43E |
1 |
A-350G/406c |
|
|
|
|
|
|
|
|
|
|
G |
|
|
c |
|
|
L83V |
|
|
|
|
|
|
|
- |
1 |
A-109c/350G |
c |
|
|
|
|
|
|
|
|
|
G |
|
|
|
|
|
L83V |
|
|
|
|
|
|
|
- |
2 |
A-257G/350G |
|
|
|
|
|
G |
|
|
|
|
G |
|
|
|
|
|
I52V L83V |
|
|
|
|
|
|
|
- |
1 |
A-257C/350G |
|
|
|
|
|
C |
|
|
|
|
G |
|
|
|
|
|
I52L L83V |
|
|
|
|
|
|
|
- |
1 |
A-257G/261G/350G/448a |
|
|
|
|
|
G |
G |
|
|
|
G |
|
|
|
a |
|
I52V V53G L83V |
|
|
|
|
|
|
|
- |
1 |
A-239A/257G/350G/368T |
|
|
|
|
A |
G |
|
|
|
|
G |
T |
|
|
|
|
A46T I52V L83V E89L |
|
|
|
|
|
|
|
- |
1 |
D |
|
|
|
T |
|
|
|
a |
g |
T |
G |
|
|
|
|
|
Q14H H78Y L83V |
|
|
|
|
c |
g |
|
- |
1 |
4.7% |
31 |
63.3 |
32 c/c |
16 c/c |
D-532g/732c |
|
|
|
T |
|
|
|
a |
g |
T |
G |
|
|
|
|
g |
Q14H H78Y L83V |
|
|
|
c |
c |
g |
|
- |
1 |
D-532g |
|
|
|
T |
|
|
|
a |
g |
T |
G |
|
|
|
|
g |
Q14H H78Y L83V |
|
|
|
|
c |
g |
|
- |
1 |
B |
|
C |
G |
T |
|
|
|
a |
g |
T |
|
|
|
|
|
|
R10T Q14D H78Y |
|
|
|
|
c |
g |
|
- |
1 |
C |
c |
T |
G |
T |
|
|
|
a |
g |
T |
|
|
g |
|
|
|
R10I Q14D H78Y |
|
G |
|
|
c |
g |
|
N29S |
1 |
The four major lineages of HPV16 were found in the studied population and E6/E7 sequences maximum likelihood tree was constructed to confirm the presence of the two principal isolates (EUR-350T prototype NC_001526.2 or EUR-350G) (Figure 1). The large majority of samples (95%) contained HPV16 sequences belonging to sublineage A1, and were identical either to the prototype EUR-350T (37.7%) or to the EUR-350G isolate (57.6%) (Table 1). The number of women infected with EUR-350T or EUR-350G during these four years showed no important variation of distribution (Figure 2). Sequence analyses of sublineage A1 showed substitutions in 10 nucleotides located between positions 109 and 856 in the E6-E7 sequence, with no changes in predicted amino acid sequences for EUR-350T isolates (2 silent mutations), and with a change for 4 EUR-350G isolates (equaling six amino acid changes I52V, L83V, I52L, V53G, A46T, E89L). The non-European HPV16 variants (lineage B, C and D) were found in five ASCUS samples (Table 1).
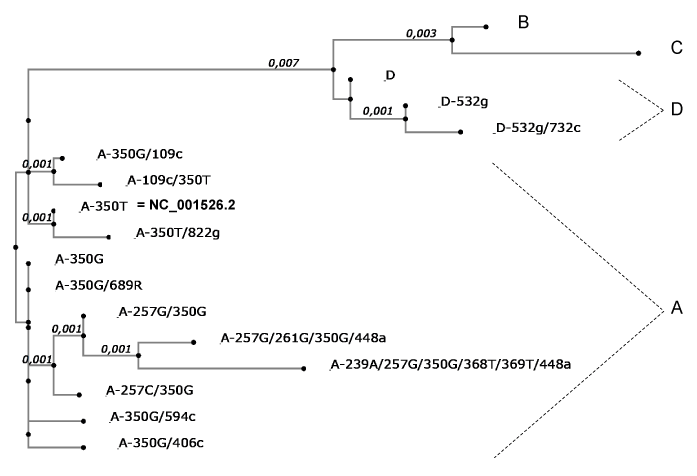
Figure 1. Phylogenetic tree of HPV16 isolates in the studied population, based on E6/E7 sequences. Each isolates are listed in table 1. The first letter is the lineage and after, letter represents the nucleotide change and number the position. Maximum likelihood tree was constructed using MAFFT software application v6.846, inferred from the global alignment of E6/E7 nucleotide sequences (nt 83-858) and referred to the prototype HPV16 sequence (NCBI Reference Sequence: NC_001526.2). The length of solid lines represents distance between isolates. Phylogenetic analysis confirmed the presence of the four lineages and the presence of the two major isolates (EUR-350T prototype NC_001526.2 or EUR-350G).
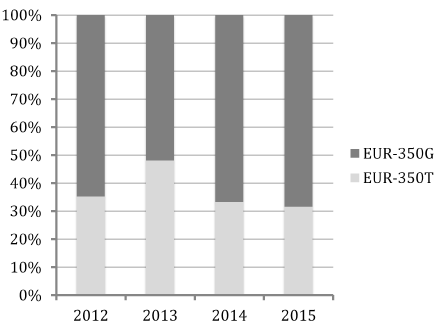
Figure 2. Ratio of isolates stratified into EUR-350T and EUR-350G during 2012–2015.
In women infected with non-European HPV16 variants (lineage B, C or D), the patient age ranged 20–55 years, with a mean age of 31.0 years. In women infected with the EUR-350T or EUR-350G variants, patient age ranged 23-57 years (with a mean age of 35.7 years) and 19-69 years (with a mean age of 35.1 years), respectively (Table 1).
HPV16 DNA status and HPV16 variants
We analyzed the physical status of HPV16 in the cervical smears that were diagnosed as ASCUS. The physical state of HPV16 was determined using qPCR that targeted the HPV16 genes E6 and E2, because the E2 gene is more frequently lost during viral integration. Thus, E2 and E6 genes are present in equivalent amounts in cells with only episomal HPV genomes, whereas in cells with 100% integrated HPV genomes, the E6 gene is present but the E2 gene is absent [38]. The percentage of integrated HPV DNA was expressed as the ratio of: the integrated viral load ([number of E6 DNA copies] - [number of E2 DNA copies] per cell) / the to number of E6 DNA copies x100. An undetectable E2 gene defined integrated forms as detected in SiHa cells control.
On the basis of the assumption that integration of HPV results in the disruption of the E2 or E1 genes, we can induce, in this analysis, an underestimate of integration. But some studies suggest that (1) the most common patterns of deletions is in the region of the E2 ORF corresponding to the protein “hinge” region [39,40], and (2) it is only the loss of regulatory episomal E2 in cells, which confers a growth advantage on selected cells [41,42].
In this study, the population infected with non-European HPV16 variants presented a high percentage of integration (mean of percentage of integration in this population = 63.3%), compared to those infected with European HPV16 variants (mean = 31.9%) with 2 samples in lineage D and 1 sample in lineage C found 100% integrated. In the population infected with sublineage A1 isolates, which were then classified into EUR-350T and EUR-350G, we observed an increased mean percentage of integration in EUR-350G (36.3%) compared to EUR-350T (25%), with significant differences in percentage of integration in these two populations infected with EUR-350T or EUR-350G: p < 0.05, as determined using the Student’s t-test (Table 1). In women infected with the EUR-350T isolate, we observed a higher number of samples without any integrated HPV genome (19 cases) than in samples of women infected with the EUR-350G isolate (5 cases) with Chi-square (χ2) test p<0.001 (5.6.10exp-6). Interestingly, box plot analysis (Figure 3) confirmed this difference of HPV16 DNA physical status between these two populations. The median of the percentage of integration was 15.70%, with 75th percentiles being 39.56% and 25th percentiles being zero, in the population infected with the EUR-350T isolate. By contrast, in the population infected with the EUR-350G isolate, the median of the percentage of integration was higher (39.80%), with a distribution of values that were more homogeneous (75th percentiles = 46.45%, and 25th percentiles = 32.39%).
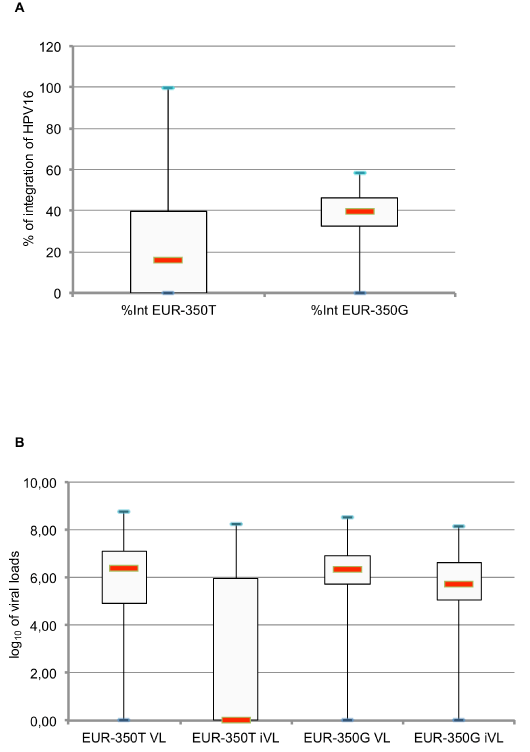
Figure 3. Distribution of HPV16 integration percentage of (A) log10 of [total HPV16 DNA viral loads per cell], and log10 of [integrated DNA HPV16 viral loads per cell] (B) in a population infected either with EUR-350T or EUR 350G
Total or integrated viral load and HPV16 variants
We analyzed the physical status of HPV16 by calculating the total viral load (number of E6 DNA copies per cell [c/c]) and the integrated viral load ([number of E6 DNA copies] - [number of E2 DNA copies] per cell), normalized for cells by quantification of β-globin DNA; in table 1, we show the means of viral load and of integrated viral load in each population. The means of the total HPV16 viral load in cervical smears from women diagnosed with ASCUS were 35 c/c and 16 c/c, respectively, in the population infected with the EUR-350T or EUR-350G HPV16 isolates. The mean of total viral load was 32 c/c in the population infected with non-European HPV16 isolates, and this population was very heterogeneous for this marker. The mean of integrated viral load was identical in the populations infected with the EUR-350T and EUR-350G variants (7 c/c and 6 c/c, respectively), and was higher in the populations infected with the non-European HPV16 variants (16 c/c). The associations between HPV16 viral loads with European or non-European HPV16 isolate parameters are shown in table 1.
The distribution of log10-transformed HPV16 viral load per million of cells (total and integrated) in the population infected with EUR-350T or EUR-350G HPV16 isolates is shown in figure 3. The log10 of total viral load per cell in cervical smears infected with the EUR-350T or EUR-350G HPV16 isolates were similar, ranging 0-8.76 c/c (median = 6.41 c/c) and 0-8.50 (median = 6.33 c/c), respectively. An analysis of distribution of log10-integrated viral load per million of cells showed a higher median in cervical smears from women infected with EUR-350G HPV16 isolates (5.70) compared to cervical smears from women infected with EUR-350T HPV16 isolates (0.00). The box plot analysis, in particular, demonstrated that the distribution of integrated viral load was more heterogeneous in the population infected with the EUR-350T isolate (75th percentiles = 5.95%, and 25th percentiles = 0.00%) than in the population infected with the EUR-350G isolate (75th percentiles = 6.62%, and 25th percentiles = 5.06%).
Discussion
Infection with high-risk Human Papillomavirus (HPV) is necessary for the development of a cervical lesion, but only a fraction of precursor lesions progress to cancer. Additional factors are likely to increase the probability of this progression occurring. For example, the percentage of HPV16 DNA integration, as a marker for high-grade cervical lesions, was demonstrated in some studies [23], while the increased frequency of non-European HPV16 variants in invasive lesions was described by Tornesello et al. [43], suggesting greater oncogenicity for the non-European HPV16 variants [44-46].
Ours is the first study on ASCUS cytology and HPV16 infection in France in which the viral lineages or isolates, plus the physical state and viral loads of HPV16 (total and integrated), have been characterized. We analyzed a population categorized by the function of the detected HPV16 isolate. Lineage A accounts for a large proportion of HPV16 in many regions and this specific ratio (EUR-350T/EUR-350G) varies considerably by world region [47]. We first analyzed the HPV16 sequences belonging to sublineage A1, which are further divided into the EUR-350T and EUR-350G isolates. The EUR-350G isolate was 1.5 times more represented than the EUR-350T isolate in this cohort of women, with stability of this ratio (EUR-350T/EUR-350G) existing for the 2012–2015 period, like in other French studies [48]. A small fraction of these lineages contained co-mutations that were previously described [49,50].
Some studies showed higher HPV16 viral loads in high-grade lesion [51]; and other studies showed integrated viral load versus total viral load is an interesting marker with which to describe cervical disease progression [52]. For HPV16 DNA, we analyzed the means for viral load and integrated viral load in cervical smears from woman diagnosed with ASCUS. The mean for HPV16 integrated viral load was identical in the two populations infected with EUR-350T or EUR-350G. Interestingly, we identified a higher median of log10-integrated viral load and even more notably, a more homogeneous distribution of log10-integrated viral load in cervical smears from women infected with EUR-350G compared to women infected with EUR-350T.
Associated with this first observation, we analyzed the physical state of HPV16 DNA in these two populations. Detection of integrated HPV might be a promising marker for cervical disease progression, because integration of HPV already occurs at an early stage of the disease [22,53]. Regarding the percentages of HPV16 DNA integration in the population infected with EUR-350G vs. with EUR-350T, we demonstrated that the mean (36.3% vs. 25.0%, respectively) and, to a greater extent, the median (39.80% vs. 15.70%, respectively) were more important in the population infected with EUR-350G (with significant differences in percentage of integration in these two populations p < 0.05, Student’s t-test) as described in study from the south of Poland [54]. Integration of HPV into the human genome often causes a lack of the E2 repressor protein, consequently inducing the up-regulation of the HPV oncogenes E6 and E7 [23,55] with an increased stability in these mRNAs [40]. The analysis of box plots showed more homogeneous values of percentage of integration in the population infected with EUR-350G vs. EUR-350T. Furthermore, we determined that there was a significant smaller number of completely episomal HPV16 genomes in this EUR-350G population. These findings suggest a lower risk of persistence and progression in women with ASCUS Pap smears that were infected with the EUR-350T isolate [8,15,20]. However our technique risks to under-quantify the percentage of integration if the gene E2 is intact after integration, for example in case of tandem-repeated HPV integrated genomes induce by the rolling circle replication mechanism [56,57]. But, HR-HPV DNA is frequently integrated into the chromosome of cervical cancer cells and is occasionally found amplified as tandem-repeated HPV integrated genomes, as typically observed in CaSki cells [21,40]. In other hand, the detection of HPV integration sites can be more relevant to the treatment monitoring of patients [58].
Furthermore, the analysis of these two markers (integrated HPV16 viral load and percentage of HPV16 DNA integration) in the few samples where a non-European HPV16 variant was found showed values that were much higher than the samples infected with a European variant. This result suggests that non-European HPV16 variants are more oncogenic than European variants, and is in agreement with the findings of previous studies [5,44,59,60] demonstrating that HPV16 non-European viral variants were significantly more likely than European variants to cause persistence and a CIN3+ evolution [8,14,16,61] inducing proliferation phenotype in an organotypic epithelial model [45 ,46], probably due to 1) modification of the immunogenic properties of the virus [46,62,63], 2) stimulation of cell migration and cell invasion [64] and 3) viral DNA integration into the host genome [45,46].
In France, the guidelines consider the test for HPV as a possible option only for managing ASCUS Pap smears. This pilot study pinpoint the potential predictive benefits of sequencing E6-E7 for the early identification of women with a higher risk of CIN3+ evolution, specifically in a HPV16-infected patient population with ASCUS Pap smears. The identification of the EUR-350G isolate (being easily detectable by short region sequencing) in women with ASCUS Pap smears should be considered as a risk indicator, and should constitute an alert signal for the medical staff [54]. This pilot study should be follow-up by a larger study to consolidate this result.
Methods
Collection of samples
Samples were obtained from women during gynecological visits in the Department of Obstetrics and Gynecology of the Archet Hospital (Nice, France) during 2012-2015. This study was carried out with approval from the local ethical committee, « Comité de Protection des Personnes » 2011-002954-28, with written informed consent from all women. Cervical smears were collected using a cytobrush. Cervical cells were placed in PreservCyt® solution (Cytyc), and were then transferred to the Pathology Laboratory for cytological screening and classification. In the University Hospital of Nice, if the results of routine Pap Tests indicated a diagnosis of ASCUS, the remainder of the sample was tested using PapilloCheck HPV DNA chips; samples positive for HPV16 were sequenced and evaluated for HPV16 physical status.
DNA isolation for PCR or HPV genotyping
For DNA isolation, 1 mL of cell suspension was centrifuged at 2000 × g for 5 min, and the cells were then resuspended in 500 µL PBS (1x). For extraction of HPV DNA, a total nucleic acid automated extraction method was used (EasyMag®, Biomérieux), according to the manufacturer’s instructions. DNA was extracted using the Generic 2.0.1 program, with onboard lysis protocol. Then, 50 µL of NucliSENS easyMAG magnetic silica was added and thoroughly mixed with the sample. Elution was performed in 25 μL NucliSens Extraction Buffer 3.
PapilloCheck® HPV DNA chips
HPV genotyping was performed using PapilloCheck® HPV DNA chips (GBO). After extraction, a 350-bp fragment of the E1 ORF, an external PCR control, and a DNA fragment of the human ADAT1 gene were amplified with specific primers (in the presence of 5 μL of each purified DNA). PCR was performed for 40 cycles at 95 °C for 30 s, 55 °C for 25 s, and 72 °C for 45 s, then 15 cycles at 95 °C for 30 s, and 72 °C for 45 s. The denatured amplification products were then hybridized to complementary DNA-probes on the chip, at room temperature, in a humid atmosphere, for 15 min. Arrays were then washed for 10 s in washing solution I, 60 s in washing solution II at 50 °C, and 10 s in washing solution III; arrays were then spun dry. Slides were scanned and analyzed using the CheckReport™ software package (GBO). The HPV genotyping results are available only if CheckReport™ software validate the ampli?cation quality of the controls (external PCR control, and a PCR of the human ADAT1 gene). The oligonucleotide microarray detects 15 HR HPV and 11 LR HPV (HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68; and HPV 6, 11, 40, 42, 43, 44/55, 53, 70, 73, 82, respectively). The PapilloCheck® DNA chip contains 28 probes, 24 HPV probes, and 4 control probes (orientation, hybridization, external PCR, and ADAT1), each in 5 replicate spots.
E6-E7 HPV sequence and phylogenetic analysis
HPV16-positive samples were further amplified with specific sets of oligonucleotides designed to amplify the ORF of E6-E7 (Table 2). PCR amplification of HPV16 E6-E7 was performed with the Invitrogen® ThermalAce DNA Polymerase kit: 3 µL of purified DNA, 5 µL of buffer 10x, 100 ng of each primer (HPV16E6vF [5′-3′] and HPV16E7vR [5′- 3′]), and 1 µL Taq polymerase. PCR was performed for 40 cycles at 95 °C for 1 min, 51 °C for 1 min, and 74 °C for 1 min. PCR products were electrophoresed in 1% agarose gel, and were visualized by staining with ethidium bromide.
Table 2. Primers and probes for quantification of E6 and E2 HPV16 DNA
Names of Primers and Probes |
SEQ 5′à3′ |
Sequence length (bp) |
Start of nucleotide position |
Amplicon length (bp) |
HPV16 E6vF |
AACCGAAACCGGTTAGTATAAA |
22 |
49 |
871 |
HPV16 E7vR |
ACCATCCATTACATCCCGTA |
20 |
900 |
16E6 F1 |
GAGAACTGCAATGTTTCAGGACC |
23 |
94 |
81 |
16E6 R1 |
TGTATAGTTGTTTGCAGCTCTGTGC |
25 |
150 |
16E6 Probe JOE |
CAGGAGCGACCCAGAAAGTTACCACAGTT |
29 |
119 |
|
16E2 F1 |
AACGAAGTATCCTCTCCTGAAATTATTAG |
29 |
3361 |
82 |
16E2 R1 |
CCAAGGCGACGGCTTTG |
17 |
3426 |
16E2 Probe FAM |
CACCCCGCCGCGACCCATA |
19 |
3406 |
|
β-globin F |
TGCATCTGACTCCTGAGGAGAA |
22 |
2379 |
81 |
β-globin R |
GGGCCTCACCACCAACTTC |
19 |
2441 |
β-glo Probe FAM |
CTGCCGTTACTGCCCT |
16 |
2403 |
|
HPV16 variants were identified by sequencing 3 µL of E6-E7 PCR product using the Big-Dye Terminator ready reaction kit (perking-Elmer), using the same primers used for initial amplification. Sequencing was run for 10 min at 96 °C, followed by 25 cycles at 96 °C for 10 s, 50 °C for 5 s, and 60 °C for 4 min. Sequencing results were analyzed (after precipitation) using an ABI PRISM 3100 genetic analyzer (Perkin-Elmer) and Sequence Navigator™ software. Only nucleotide changes that were verified as occurring on both strands were accepted.
The variants were identified and numbered using the prototype sequence (HPV16, NCBI Reference Sequence: NC_001526.2) belonging to the European lineage. Sequence alignments and multiple alignments of E6-E7 were performed using the BioEdit Sequence Alignment Editor, and the phylogenetic tree was constructed using MAFFT software application, v6.846.
Quantitative real time-PCR (qPCR)
PCR amplifications were performed on an Applied Biosystems StepOne Plus Real-Time PCR system, using TaqMan Fast Universal Master Mix (Life Technologies). The amplification conditions were 95 °C for 20 s, 95°C for 1 s, and 60 °C for 20 s, for 40 cycles. The primers and probes were described previously [22] and real-time PCR was performed for a simultaneous amplification of two HPV16 genes: E2 and E6 (Table 2). Each sample was normalized using β-globin in separate reactions; the β-globin gene was used to correct HPV copy numbers for cells in the cervical swab material. The final primer and probe concentrations, in a total volume of 10 µL, were 0.9 µM and 0.25 µM, respectively. A volume of 2.5 µL of target DNA from cervical smears was added to the reaction mixture.
Three standard curves were obtained by amplification of a serial dilution of 70 million to 700 copies of the HPV16 E2 and E6 plasmids (Taq-amplified PCR E2 or E6 product into a plasmid vector in pCR™2.1 Vector) and the β-globin plasmid (Taq-amplified PCR β-globin product into a plasmid vector in pCR™2.1 Vector). There was a linear relationship between the threshold cycle values plotted against the log of the copy number, over the entire range of dilutions. HPV16 E6 and E2 viral loads were expressed as the number of HPV16 copies per cell equivalent. The integration of HPV16 into human genome was detected by subtracting the copy numbers of E2 (episomal DNA) from the total copy numbers of E6 (episomal and integrated DNA combined).
Different physical states (episomal, integrated or both) were identified by the percentage of integrated HPV DNA was expressed as the ratio of the integrated viral load to the total copy numbers of E6. SiHa cells, which represent cells containing fully integrated HPV, were included in each run to determine the detection limits for integrated HPV16. All experiments were performed in duplicate, with similar ratios. We calculated the percentage of integrated HPV DNA of each samples and the mean of this percentage in each lineages. An undetectable E2 threshold cycle defined integrated forms as detected in SiHa cells control.
Statistical analysis
The significance of number of samples without any integrated HPV genome between HPV16 sublineages groups was tested using the chi-square method. Differences in viral quantitative data associated in population infected with different HPV16 sublineages were compared using the Student’s t-test. All p values presented were interpreted as significant when p < 0.05.
Statistical analyses were conducted using the Easy Med Stat© software package.
Funding information
This work was supported by institutional grants from CHU and Conseil Général 06.
Acknowledgments
This work was supported by institutional grants from CHU de Nice. Professor Michiels is kindly acknowledged for instruction and for valuable advice. We thank Ms. Forrest and EditageOnlineTM for polishing the English form of the manuscript.
2021 Copyright OAT. All rights reserv
ethical statement
This study was carried out with approval from the local ethical committee, « Comité de Protection des Personnes » 2011-002954-28. Findings will be reported in scientific publications and at research conferences.
References
- Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, et al. (1999) Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol 189: 12-19. [Crossref]
- de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H (2004) Classification of papillomaviruses. Virology 324: 17-27. [Crossref]
- Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, et al. (2003) Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med 348: 518-527. [Crossref]
- Clifford GM, Smith JS, Plummer M, Muñoz N, Franceschi S (2003) Human papillomavirus types in invasive cervical cancer worldwide: a meta-analysis. Br J Cancer 88: 63-73. [Crossref]
- Sichero L, Ferreira S, Trottier H, Duarte-Franco E, Ferenczy A, et al. (2007) High grade cervical lesions are caused preferentially by non-European variants of HPVs 16 and 18. Int J Cancer 120: 1763-1768. [Crossref]
- Badano I, Totaro ME, Culasso AC, Sanabria DJ, Schurr TG, et al. (2015) Genetic characterization and clinical implications of human papillomavirus type 16 (HPV16) variants from northeastern Argentina. Infect Genet Evol 29:103-109. [Crossref]
- Sun M, Gao L, Liu Y, Zhao Y, Wang X, et al. (2012) Whole genome sequencing and evolutionary analysis of human papillomavirus type 16 in central China. PLoS One 7: e36577. [Crossref]
- Grodzki M, Besson G, Clavel C, Arslan A, Franceschi S, et al. (2006) Increased risk for cervical disease progression of French women infected with the human papillomavirus type 16 E6-350G variant. Cancer Epidemiol Biomarkers Prev 15: 820-822. [Crossref]
- Mirabello L, Yeager M, Cullen M, Boland JF, Chen Z, et al. (2016) HPV16 Sublineage Associations with Histology-Specific Cancer Risk Using HPV Whole-Genome Sequences in 3200 Women. J Natl Cancer Inst 108. [Crossref]
- Bernard HU, Burk RD, Chen Z, van Doorslaer K, zur Hausen H, et al. (2010) Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 401: 70-79. [Crossref]
- Cornet I, Gheit T, Franceschi S, Vignat J, Burk RD, et al. (2012) Human papillomavirus type 16 genetic variants: phylogeny and classification based on E6 and LCR. J Virol 86: 6855-6861. [Crossref]
- Burk RD, Harari A, Chen Z (2013) Human papillomavirus genome variants. Virology 445: 232-243. [Crossref]
- Yamada T, Manos MM, Peto J, Greer CE, Munoz N, et al. (1997) Human papillomavirus type 16 sequence variation in cervical cancers: a worldwide perspective. J Virol 71: 2463-2472. [Crossref]
- Schiffman M, Rodriguez AC, Chen Z, Wacholder S, Herrero R, et al. (2010) A population-based prospective study of carcinogenic human papillomavirus variant lineages, viral persistence, and cervical neoplasia. Cancer Res 70: 3159-3169. [Crossref]
- Zhang L, Yang B, Zhang A, Zhou A, Yuan J, et al. (2016) Association between human papillomavirus type 16 E6 and E7 variants with subsequent persistent infection and recurrence of cervical high-grade squamous intraepithelial lesion after conization. J Med Virol 88: 1982-1988. [Crossref]
- Zehbe I, Wilander E, Delius H, Tommasino M (1998) Human papillomavirus 16 E6 variants are more prevalent in invasive cervical carcinoma than the prototype. Cancer Res 58: 829-833. [Crossref]
- Da Costa MM, Hogeboom CJ, Holly EA, Palefsky JM (2002) Increased risk of high-grade anal neoplasia associated with a human papillomavirus type 16 E6 sequence variant. J Infect Dis 185: 1229-1237. [Crossref]
- Wu Y, Chen Y, Li L, Yu G, He Y, et al. (2006) Analysis of mutations in the E6/E7 oncogenes and L1 gene of human papillomavirus 16 cervical cancer isolates from China. J Gen Virol 87: 1181-1188.
- Xi LF, Koutsky LA, Hildesheim A, Galloway DA, Wheeler CM, et al. (2007) Risk for high-grade cervical intraepithelial neoplasia associated with variants of human papillomavirus types 16 and 18. Cancer Epidemiol Biomarkers Prev 16: 4-10. [Crossref]
- Lee K, Magalhaes I, Clavel C, Briolat J, Birembaut P, et al. (2008) Human papillomavirus 16 E6, L1, L2 and E2 gene variants in cervical lesion progression. Virus Res 131: 106-110. [Crossref]
- Pett M, Coleman N (2007) Integration of high-risk human papillomavirus: a key event in cervical carcinogenesis? J Pathol 212: 356-367. [Crossref]
- Peitsaro P, Johansson B, Syrjanen S (2002) Integrated human papillomavirus type 16 is frequently found in cervical cancer precursors as demonstrated by a novel quantitative real-time PCR technique. J Clin Microbiol 40: 886-891. [Crossref]
- Cricca M, Morselli-Labate AM, Venturoli S, Ambretti S, Gentilomi GA, et al. (2007) Viral DNA load, physical status and E2/E6 ratio as markers to grade HPV16 positive women for high-grade cervical lesions. Gynecol Oncol 106: 549-557. [Crossref]
- Theelen W, Speel EJ, Herfs M, Reijans M, Simons G, et al. (2010) Increase in viral load, viral integration, and gain of telomerase genes during uterine cervical carcinogenesis can be simultaneously assessed by the HPV 16/18 MLPA-assay. Am J Pathol 177: 2022-2033. [Crossref]
- Wentzensen N, Vinokurova S, von Knebel Doeberitz M (2004) Systematic review of genomic integration sites of human papillomavirus genomes in epithelial dysplasia and invasive cancer of the female lower genital tract. Cancer Res 64: 3878-3884. [Crossref]
- Aromseree S, Chaiwongkot A, Ekalaksananan T, Kongyingyoes B, Patarapadungkit N, et al. (2014) The three most common human papillomavirus oncogenic types and their integration state in Thai women with cervical precancerous lesions and carcinomas. J Med vir 86: 1911-1919. [Crossref]
- Das P, Thomas A, Kannan S, Deodhar K, Shrivastava SK, et al. (2015) Human papillomavirus (HPV) genome status & cervical cancer outcome--A retrospective study. Indian J Med Res 142: 525-532. [Crossref]
- Manawapat A, Stubenrauch F, Russ R, Munk C, Kjaer SK, et al. (2012) Physical state and viral load as predictive biomarkersfor persistence and progression of HPV16-positive cervical lesions: results from a population based long-term prospective cohort study. Am J Cancer Res 2: 192-203. [Crossref]
- Sun CA, Lai HC, Chang CC, Neih S, Yu CP, et al. (2001) The significance of human papillomavirus viral load in prediction of histologic severity and size of squamous intraepithelial lesions of uterine cervix. Gynecol Oncol 83: 95-99. [Crossref]
- Sun CA, Liu JF, Wu DM, Nieh S, Yu CP, et al. (2002) Viral load of high-risk human papillomavirus in cervical squamous intraepithelial lesions. Int J Gynaecol Obstet 76: 41-47. [Crossref]
- Gravitt PE, Kovacic MB, Herrero R, Schiffman M, Bratti C, et al. (2007) High load for most high risk human papillomavirus genotypes is associated with prevalent cervical cancer precursors but only HPV16 load predicts the development of incident disease. Int J Cancer 121: 2787-2793. [Crossref]
- Moberg M, Gustavsson I, Wilander E, Gyllensten U (2005) High viral loads of human papillomavirus predict risk of invasive cervical carcinoma. Br J Cancer 92: 891-894. [Crossref]
- Lorincz AT, Castle PE, Sherman ME, Scott DR, Glass AG, et al. (2002) Viral load of human papillomavirus and risk of CIN3 or cervical cancer. Lancet 360: 228-229. [Crossref]
- Castle PE, Schiffman M, Scott DR, Sherman ME, Glass AG, et al. (2005) Semiquantitative human papillomavirus type 16 viral load and the prospective risk of cervical precancer and cancer. Cancer Epidemiol Biomarkers Prev 14: 1311-1314. [Crossref]
- Carcopino X, Henry M, Benmoura D, Fallabregues AS, Richet H, et al. (2006) Determination of HPV type 16 and 18 viral load in cervical smears of women referred to colposcopy. J Med Virol 78: 1131-1140. [Crossref]
- Constandinou-Williams C, Collins SI, Roberts S, Young LS, Woodman CB, et al. (2010) Is human papillomavirus viral load a clinically useful predictive marker? A longitudinal study. Cancer Epidemiol Biomarkers Prev 19: 832-837. [Crossref]
- Seedorf K, Krämmer G, Dürst M, Suhai S, Röwekamp WG (1985) Human papillomavirus type 16 DNA sequence. Virology 145: 181-185. [Crossref]
- Cañadas MP, Darwich L, Sirera G, Cirigliano V, Bofill M, et al. (2010) New molecular method for the detection of human papillomavirus type 16 integration. Clin Microbiol Infect 16: 836-842. [Crossref]
- Kalantari M, Karlsen F, Kristensen G, Holm R, Hagmar B, et al. (1998) Disruption of the E1 and E2 reading frames of HPV 16 in cervical carcinoma is associated with poor prognosis. Int J Gynecol Pathol 17: 146-153. [Crossref]
- Jeon S, Lambert PF (1995) Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proc Natl Acad Sci U S A 92: 1654-1658. [Crossref]
- Pett MR, Herdman MT, Palmer RD, Yeo GS, Shivji MK, et al. (2006) Selection of cervical keratinocytes containing integrated HPV16 associates with episome loss and an endogenous antiviral response. Proc Natl Acad Sci U S A 103: 3822-3827. [Crossref]
- Sang BC, Barbosa MS (1992) Increased E6/E7 transcription in HPV 18-immortalized human keratinocytes results from inactivation of E2 and additional cellular events. Virology 189: 448-455. [Crossref]
- Tornesello ML, Duraturo ML, Salatiello I, Buonaguro L, Losito S, et al. (2004) Analysis of human papillomavirus type-16 variants in Italian women with cervical intraepithelial neoplasia and cervical cancer. J Med Vir 74: 117-126. [Crossref]
- Freitas LB, Chen Z, Muqui EF, Boldrini NA, Miranda AE, et al. (2014) Human papillomavirus 16 non-European variants are preferentially associated with high-grade cervical lesions. PloS one 9: e100746. [Crossref]
- Jackson R, Rosa BA, Lameiras S, Cuninghame S, Bernard J, et al. (2016) Functional variants of human papillomavirus type 16 demonstrate host genome integration and transcriptional alterations corresponding to their unique cancer epidemiology. BMC Genomics 17: 851. [Crossref]
- Jackson R, Togtema M, Lambert PF, Zehbe I (2014) Tumourigenesis driven by the human papillomavirus type 16 Asian-American e6 variant in a three-dimensional keratinocyte model. PloS one 9: e101540. [Crossref]
- Cornet I, Gheit T, Iannacone MR, Vignat J, Sylla BS, et al. (2013) HPV16 genetic variation and the development of cervical cancer worldwide. Br J Cancer 108: 240-244. [Crossref]
- Cornet I, Gheit T, Clifford GM, Combes JD, Dalstein V, et al. (2013) Human papillomavirus type 16 E6 variants in France and risk of viral persistence. Infect Agent Cancer 8: 4.
- de Boer MA, Peters LA, Aziz MF, Siregar B, Cornain S, et al. (2004) Human papillomavirus type 16 E6, E7, and L1 variants in cervical cancer in Indonesia, Suriname, and The Netherlands. Gynecol Oncol 94: 488-494. [Crossref]
- Ortiz-Ortiz J, Alarcon-Romero Ldel C, Jimenez-Lopez MA, Garzon-Barrientos VH, Calleja-Macias I, et al. (2015) Association of human papillomavirus 16 E6 variants with cervical carcinoma and precursor lesions in women from Southern Mexico. Vir J 12: 29. [Crossref]
- Saunier M, Monnier-Benoit S, Mauny F, Dalstein V, Briolat J, et al. (2008) Analysis of human papillomavirus type 16 (HPV16) DNA load and physical state for identification of HPV16-infected women with high-grade lesions or cervical carcinoma. J Clin Microbiol 46: 3678-3685. [Crossref]
- Fontaine J, Gravitt P, Duh LM, Lefevre J, Pourreaux K, et al. (2005) High level of correlation of human papillomavirus-16 DNA viral load estimates generated by three real-time PCR assays applied on genital specimens. Cancer Epidemiol Biomarkers Prev 14: 2200-2207. [Crossref]
- Collins SI, Constandinou-Williams C, Wen K, Young LS, Roberts S, et al. (2009) Disruption of the E2 gene is a common and early event in the natural history of cervical human papillomavirus infection: a longitudinal cohort study. Cancer Res 69: 3828-3832. [Crossref]
- Szostek S, Zawilinska B, Klimek M, Kosz-Vnenchak M (2017) HPV16 E6 polymorphism and physical state of viral genome in relation to the risk of cervical cancer in women from the south of Poland. Acta Biochim Pol 64: 143-149. [Crossref]
- Raybould R, Fiander A, Hibbitts S (2011) Human Papillomavirus Integration and its Role in Cervical Malignant Progression. The Open Clinical Cancer Journal 5: 1-7.
- Kusumoto-Matsuo R1, Kanda T, Kukimoto I (2011) Rolling circle replication of human papillomavirus type 16 DNA in epithelial cell extracts. Genes Cells 16: 23-33. [Crossref]
- Xu F, Cao M, Shi Q, Chen H, Wang Y, et al. (2015) Integration of the full-length HPV16 genome in cervical cancer and Caski and Siha cell lines and the possible ways of HPV integration. Virus Genes 50: 210-220. [Crossref]
- Xu B, Chotewutmontri S, Wolf S, Klos U, Schmitz M, et al. (2013) Multiplex Identification of Human Papillomavirus 16 DNA Integration Sites in Cervical Carcinomas. PLoS One 8: e66693. [Crossref]
- Xi LF, Koutsky LA, Galloway DA, Kuypers J, Hughes JP, et al. (1997) Genomic variation of human papillomavirus type 16 and risk for high grade cervical intraepithelial neoplasia. J Natl Cancer Inst 89: 796-802. [Crossref]
- Cheung JL1, Lo KW, Cheung TH, Tang JW, Chan PK (2006) Viral load, E2 gene disruption status, and lineage of human papillomavirus type 16 infection in cervical neoplasia. J Infect Dis 194: 1706-1712. [Crossref]
- Xi LF, Demers GW, Koutsky LA, Kiviat NB, Kuypers J, et al. (1995) Analysis of human papillomavirus type 16 variants indicates establishment of persistent infection. J Infect Dis 172: 747-755. [Crossref]
- Kumar A, Hussain S, Yadav IS, Gissmann L, Natarajan K, et al. (2015) Identification of human papillomavirus-16 E6 variation in cervical cancer and their impact on T and B cell epitopes. J Virol Methods 218: 51-58. [Crossref]
- Song D, Li H, Li H, Dai J (2015) Effect of human papillomavirus infection on the immune system and its role in the course of cervical cancer. Oncol Lett 10: 600-606. [Crossref]
- Hochmann J, Sobrinho JS, Villa LL, Sichero L (2016) The Asian-American variant of human papillomavirus type 16 exhibits higher activation of MAPK and PI3K/AKT signaling pathways, transformation, migration and invasion of primary human keratinocytes. Virology 492: 145-154. [Crossref]



