Chronic lymphocytic leukemia (CLL) is the most common type of leukemia among adults. It exhibits heterogeneity in its clinical course. Its cytogenetic findings play significant role in predicting treatment response and prognosis. The study was aimed to evaluate the frequencies of deletions 13q14.3, 11q22.3, trisomy 12, 17p13.1 and their role in disease progression with FISH technique. Results revealed the higher prevalence of del 13q14.3 followed by del 11q22.3, trisomy-12 and del 17p13.1. The higher frequencies of del 13q14.3 could be primary event in inducing CLL and other deletions could be drivers for CLL progression.
chronic lymphocytic leukemia, prevalence, cytogenetic, fluorescence insitu hybridization
Chronic lymphocytic leukemia (CLL) is a clonal accumulation of B lymphocytes in peripheral blood, bone-marrow and lymph nodes. CLL is the most common leukemia among adults [1]. However, in Western countries CLL is most common and reported dramatic increase in the incidence at the age more than 60 years. The male: female ratio in patients was found as 1.5-2:1. Further current data suggest the incidence of CLL as 3.5/100,000 in Europe and North America and the frequency is approximately one of every three leukemia cases [2].
The prevalence of specific abnormalities in B-CLL is currently being reassessed by FISH interphase cytogenetic. The most frequent abnormalities are deletions involving chromosome band 13q14 followed by deletions of genomic region11q22.3, trisomy-12 and mutation of TP53 tumour suppressor gene at 17p13 [3]. For decades, CLL received little attention largely because it has prolonged course, many persons affected by CLL live relatively normal life and eventually die of cause unrelated to CLL [4].
The eighty percent with CLL carry one of four common chromosomal aberrations which are detected by fluorescence insitu hybridization (FISH) analysis. The del 13q14 is the most common chromosomal aberrations in more than 50% of CLL patients. The function of deleted gene is down regulation of anti-apoptotic gene BCL-2 resistance which is associated with favourable prognosis. The deletion of 11q22.3 found in 10-20% of patients and is associated with alteration in ATM gene, which encodes proteins involve in DNA repair. Trisomy-12 was reported 15-20% in CLL which is associated with fundamental up regulation of integrin signalling. The del 17p13 is predictive worst prognosis which is found in 7-10% patients.
In the present study, we report the prevalence of cytogenetic abnormalities in CLL from the region of Western India. A total of 70 patients, suspected for CLL were screened. Of which 50 were CLL positive with age ranging from 36 to 78 years. FISH analysis revealed that out of 70 patients, at least 50 patients found with one chromosomal abnormalities which includes del 13q14, 11q22 (ATM), trisomy-12 and del 17p13(TP53).
A total of 70 heparinized bone-marrow samples of suspected CLL patents were collected for conventional cytogenetic technique. Of which 50 samples were found with CLL and 20 samples were normal. The cultures were set up, harvested and slides were prepared. The GTG banding was performed. The images of metaphases were captured and karyotype was prepared using IKAROS software. Chromosome identification and karyotyping were made according to ISCN [5]. The FISH analysis of samples was performed and chromosomal abnormalities were evaluated using CLL locus specific probes. The examined aberrations included Deletions 13q, 11q, trisomy-12 and 17p.
A total of 70 suspected subjects were enrolled, of which 50 were confirmed CLL patients. FISH analysis revealed that the rate of abnormalities were 50% for del 13q14.3 (Figure 1a), 22% for 11q22.3 (Figure 1b), 18% for trisomy-12 (Figure 1c) and 10% for del 17p13 (Figure 1d). The most noted chromosomal aberrations were del 13q followed by 11q, trisomy-12 and del 17p (Table 1).
Table1. Patients age and sex wise distribution and clinical characteristics
Age (years) |
36 –78 |
Male |
33(66%) |
Female |
17(34%) |
Chromosomal aberration |
|
13q14.3 deletion |
25(50%) |
11q22.3 deletion |
11(22%) |
Trisomy 12 |
09(18%) |
17p13.1 deletion |
05(10%) |
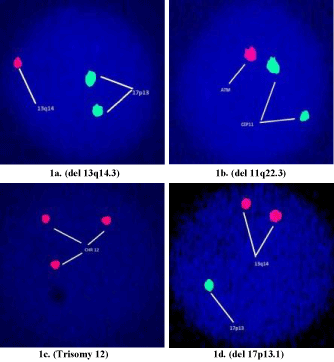
Figure 1. Detection of cytogenetic abnormalities in CLL using fluoresence in situ hybridiztion technique
During last 10 years, the understanding of CLL biology especially with genetic and molecular factors was useful for prognosis of CLL patients [6]. It is known that conventional cytogenetic analyses underestimate chromosomal rearrangements in CLL. However, FISH considered much more sensitive for detection of genomic aberrations [7]. The prevalence of specific aberrations in B-CLL is generally detected by interphase FISH technique. The most frequent abnormalities are del 13q14 followed by del 11q22.3, trisomy-12 and deletion/mutation of TP53 tumor suppressor gene at 17p13.
The FISH technique is widely used to detect genetic abnormalities because of its sensitivity and high resolution [8]. We used this technique to identify chromosomal rearrangements in CLL patients. Our results revealed that del 13q14.3 (50%) was most frequent observed abnormality. Our results are in agreements other previous studies, further second important detected was del 11q22.3 (22%) followed by trisomy-12 (18%) and TP53 mutation involving 17p13 (10%) [9].
Major geographic differences in the epidemiology of CLL between Eastern and Western countries. It has also been reported that CLL is common predominantly in European descent but rare in Asian. The pattern of chromosomal aberrations in CLL is also different in Asian and European [10]. In a recent study, it has been reported that the CLL chromosomal aberrations were del 13q followed by del 17p, 11q and trisomy-12. In our study, we observed CLL chromosomal aberrations in sequence of deletions 13q followed by 11q, triosomy-12 and 17p. Our results are not in agreement with that of Bagir, et al. [11] which proves that prognostic factors, mainly cytogenetic aberrations found in CLL patients differed between geographic regions.
Chronic lymphocytic leukemia (
CLL
) is the most common type of leukemia among adults. Our results revealed that the frequencies of deletions were in pattern of 13q14.3 followed by 11q22.3, trisomy-12 and 17p13 with most sensitive interphase FISH technique. The higher frequency of 13q14.3 could be primary event in inducing CLL and other deletions could be drivers for CLL progression.
- Donher H, Stilogenbauer S, James MA, Benner A, Weiluni, et al. (1997) 11q deletion identify a new subset of B cell chronic lymphocytic leukemia characterised by extensive nodal involvement of and interior prognosis. Blood 89: 2516-2522. [Crossref]
- Talab F, Allen JC, Thompson V, Lik K, Slupsky JR (2013) LCK is an important mediator B cell receptor signalling in chronic lymphocytic leukemia. Mol Can Res 11: 541-554. [Crossref]
- Parker H, Rose-Zerill MJ, Parker A, Chaplin T, Wade R, et al. (2011) 13q deletion anatomy and disease progression in patients with chronic lymphocytic leukemia. Leukemia 25: 489-497. [Crossref]
- Lad DP, Malhotra P, Verma S (2013) Chronic lymphocytic leukemia: Inception to cure: Are we there? Indian J Hematol Blood Transfus 29: 1-10.
- International System for Human Cytogenetics Nomenclature (ISCN), Karger Publ. Inc. (2009).
- Gribben JG, O’Brine S (2011) Update on therapy of chronic lymphocytic leukemia. J Clin Oncol 29: 544-550. [Crossref]
- Gribben JG (2010) How I treat Chronic lymphocytic leukemia up front. Blood 115: 187-197. [Crossref]
- Reddy KS (2006) Chronic lymphocytic leukemia profiled for prognosis using a fluorescence in situ hybridization panel. Br J Hematol 132: 705-722. [Crossref]
- Ripolles L, Ortega M, Ortuno F, Gonzlez A, Losada, et al. (2006) Genetic abnormalities and classical outcome in CLL. Can Genet Cyto 171: 57-64.
- Yang SM, Li JY, Gale RP, Huang XJ (2015) The mystery of chronic lymphoctic leukemia (CLL): why is it absent in Asians and what does this tell us about etiology, pathogenesis and biology? Blood Rev 29: 205-213. [Crossref]
- Bagir EK, Acikalin A, Alsancak P, Pyadas S, Gurkan E, et al. (2018) Prevalence of cytogenetic abnormalities in chronic lymphocytic leukemia in the Southern part of Turkey. Ind J Cancer 54: 573-575. [Crossref]

