Abstract
Objectives: The multiplex ligation-dependent probe amplification (MLPA) is fast, costeffective method that widely using in the prenatal screening. In the current study it was aimed to analyze the amnion and aborted materials by QF-PCR and MLPA techniques.
Materials and Methods: Total genomic DNA from 67 amnion and 11 aborted materials were identified by QF-PCR and MLPA techniques and compared. We used two distinct MLPA kits, one for aneuploidy with 36 probes and other one for microdeletion/duplications with 50 probes.
Results: The QF-PCR analysis of amnion samples were negative in 58 and positive in 9 cases, while the same analysis was negative in 2 and positive in 9 samples of aborted material, which was statistically different between two groups (p < 0.001). MLPA aneyploidi genotyped showed 57 negative and 10 positive results in amnion group, while in aborted material group this analysis was positive in all 11 cases (p < 0.001). MLPA microdeletion/duplication analysis was performed in 33 cases of amnion where results were negative in 11 and positive in 22 samples, while in aborted material this analysis was positive in all 11 samples. Microdeletions were detected in chromosomes 18 and 21 in two different samples that diagnosed as normal by QF-PCR technique in the current results. The rest of other samples were diagnosed as the same as QF-PCR technique.
Conclusion: In conclusion, the current results confirmed that both MLPA kits can be used for the prenatal diagnosis successfully, but it is better used it in combination with other techniques for prenatal diagnosis.
Key Words
Prenatal diagnosis, amnion and aborted materials, QF-PCR, MLPA techniques
Introduction
Spontaneous pregnancy loss is relatively common and occurs in about 10-15% of all clinically recognized pregnancies resulting in pregnancy failure [1]. Many factors can have influenced successful pregnancy, but around of 50% of first-trimestar abortions and one-third of second-trimester abortions are due to cytogenetic abnormalities. The most frequent cytogenetic abnormalities are numerical aberrations, mainly trisomy, polyploidy and monosomy, followed by structural rearrangements and other abnormalities such as chromosomal mosaicism [2, 3]. Today, a lot of different prenatal diagnosis techniques has aims to detect the possible trisomic and/or monosomic chromosomal abnormalities in embryo and fetal cells before birth, but also in aborted fetal material [1-5]. Over the last few decades, prenatal diagnosis of fetal chromosomal abnormalities has relied on conventional cytogenetic analysis of cultured amniocytes, chronic villi or fetal blood as a gold standard for detections chromosomal abnormalities. In the last years, alternative methods have been development to reduce reporting time, the work load and to allow the introduction of automatic methods. Molecular techniques could play a role as an additional technique when culture failure or maternal contamination occurs in standard cytogenetic technique, but also could be very important in detecting submicroscopic chromosomes variants [6]. Three rapid aneuploidy test (RATs) are used to detect the most common aneuploidies (trisomies 13, 18, 21 and sex chromosomes): fluorescence in sity hybridization, quantitative fluorescent PCR (QF-PCR) and multiplex ligation-dependent probe amplification [7]. The clinical validity of QF-PCR to detect the common aneuploidies has been reported by a number of investigators. This technique has advantage of providing rapid results for the diagnosis or exclusion of aneuploidy in chromosomes 13, 18, 21 or XY. In comparison with standard cytogenetic analysis of prenatal samples, the QF-PCR is providing to be reliable method for detection of trisomies that should replace the conventional cytogenetic analysis whenever prenatal testing is performed solely because of an increased risk of aneuploidy in chromosomes 13, 18, 21 and X or Y [7]. Both conventional and QF-PCR technic should be performed in all cases of prenatal diagnosis referred for a fetal ultrasound abnormality or a family chromosomal rearrangement [8]. Although, the application of QF-PCR as alone approach in prenatal diagnosis is much debated. The major criticism is that this test detects only numerical abnormalities of the tested chromosomes. Nevertheless, the low frequency of severe abnormalities not detected by RAT techniques but revealed by conventional karyotyping has been noted. Moreover, it has been questioned which prenatal samples should be studied only by RAT methods and which by karyotyping (with or without the RAT method) [7]. Multiple Ligation-Dependent Probe Amplification (MLPA) is a relatively new method of molecular diagnosis. It enables a relative quantitative assessment of up to many different PCR amplicons in one reaction by the use of a very small amount of examined DNA. Nowdays, MLPA is becoming a very helpful tool in prenatal diagnosis and is widely used for detection of aneuploidies, familial single gene disorders, common microdeletion syndrome and identification of marker chromosomes in fetus [9, 10]. MLPA technique offers advantage over karyotype in terms of lower failure rate, faster turnaround time and much higher resolution than conventional karyotyping [11]. Additionaly, MLPA analysis is today very powerful tool for the detection of gene deletions/duplications. This technique is able to analyse in a single reaction up to 50 DNA sequences and to detect also copy number variations of specific genes, including small intragenic rearrangements. Due to this ability, MLPA can be used in the molecular diagnosis of several genetic diseases whose pathogenesis is related to the presence of deletions or duplications of specific genes [12].
Objectives
In the current project it was aimed to check the possible advantage of the multiplex ligation-dependent probe amplification (MLPA) technique in prenatal diagnosis. Except the amnio fluid, in our study we also examined the fetal aborted material. MLPA SALSA (P095A3 for aneuploidy and P245-B1 for microdeletion/duplication) probes were used to detect cytogenetic abnormalities in amnion and production of spontaneous abortions and compared it with results obtained by the QF-PCR.
Material and Methods
This study was conducted according to the Declaration of Helsinki and the protocol was reviewed and approved by the institutional review ethics committee. It is included 67 samples of amnion and 11 samples of aborted fetal material. The reasons for prenatal amnion diagnosis were: advanced maternal age (>35 years) abnormal biochemical screening, increased nuchal translucency, maternal anxiety or other reason (mainly abnormal sonograpcic finding or family history of chromosomal rearrangements).
The DNA was isolated by commercial kit for DNA isolation, following manufactures’s protocol. We used Qiaqen, QIAmp DNA FFPE Tissue Kit for abortion material (Qiagen, GmbH, Hilden, Germany) and InstaGenetx Matrix kit (BIO RAD, USA) for amnion sample material. Molecular genetic analysis was performed by QF-PCR test using a set of STR markers for chromosomes 13, 18, 21, X and Y. All forward primers were fluorescently labelled with different fluorochromes. PCR products were analysed with an ABI3130 genetic analyser (Applied Biosystems, Foster City, CA) and Gene Mapper v3.5 software was used to analyse the results.
Two different MLPA tests were used in this study, one was SALSA P095-A3 with 36 probes for aneuploidy, and the second one was P245-B1 with 50 probes (MRC Holland) for microdeletion/duplication. MPLA involved six processes: DNA denaturation and
hybridization, ligation, PCR amplification, capillary electrophoresis and data normalization. The MLPA fragments were separated by capillary electrophoresis and the resulting probe peak areas were analysed using Coffalyser software which automatically normalizes and compares samples with the reference data. Peak height ration >1.3 interpreted as copu number gain, while peak height ration <0.7 was interpreted as copy number loss. Copy number gains or losses involving one chromosomal arm were interpreted as segmental aneyploidy, while copy number gain on both arms of chromosomes was interpreted as trisomy.
After obtaining the results we performed χ2 test in SPSS software (v.17.0 IBS SPSS Inc., SAD) to analyse the distribution between samples of amnion and fetal aborted materials. Also, we want to compere concordance between the results obtained by QF-PCR and MLPA technique.
Results
The QF-PCR analysis of amnion samples were negative in 58 (86.57%) and positive in 9 (13.43%) samples of amnion, while the same analysis was negative in 2 (18.18%) and positive in 9 (81.82%) samples of aborted material, which was statistically different between two groups (p < 0.001) (Table 1).
Table 1. The results of QF-PCR and MLPA genotyping for aneyploidy in amnion and aborted material
|
QF-PCR |
MLPA PO95 |
Material type (n=78) |
Amnion
(n=67) |
Aborted
(n=11) |
Amnion
(n=67) |
Aborted
(n=11) |
Chromosome
Profiles n
(%) |
Normal |
58 (86.6) |
2 (18.2) |
57 (85.0) |
ND |
Mutated |
Numerical |
9 (13.4) |
9 (81.8) |
10 (15) |
11 (100) |
Structural |
ND |
ND |
ND |
ND |
Χ2 test |
p |
<0.0001* |
<0.0001* |
* p < 0.05; ND-not detected
MLPA SALSA P095-A3 (with 36 probes for aneuploidy) analysis showed 57 (85.07%) negative and 10 (14.93%) positive results in amnion group, while in aborted material group this analysis was positive in all 11 cases (100%) (p < 0.001) (Table 1).
MLPA SALSA P245-B1 (with 50 probes for microdeletion/duplication) analysis was performed in 33 cases of amnion where results were negative in 11 (33.3%) and positive in 22
(66.7%) samples. The same analysis was positive in all 11 samples of aborted material (Table 2). Microdeletions were detected in chromosomes 18 and 21 in two different aborted material samples that diagnosed as normal by QF-PCR technique in the current results. Two regional chromosomal duplication were detected in chromosome 18 (18q11.2) and 21 (21q22.3) of different aborted samples in the current study. Table 3 shows all 50 analysed loci in aborted fetal material, where dark shades areas show relatice probe signal of >1.3 which was considered as duplication/partial duplication (Table 3). The light shades areas show relative probe signals of < 0.7 which was considered as deletion/partial deletion. Fig. 1 showes the normal microdeletions/duplications pik profile from the female healthy individual as a control, while Fig.2 and Fiq.3 represents the aborted material samples with heterozygous duplication (red arrows) and heterozygous deletion (blue arrows) (figure 1) (figure 2) (figure 3).
Table 2. MLPA genotyping for microdeletion/duplication for the presented amnion and aborted samples
|
Technique |
MLPA P245 |
|
2021 Copyright OAT. All rights reserv
|
Material type |
Amnion (n=33) |
Aborted (n=11) |
|
|
Gender |
M
(n=15) |
F (n=18) |
M
(n=5) |
F (n=6) |
p |
Chromosome
Profiles n (%) |
Normal |
19 (57.6) |
ND |
0.0001* |
Mutated |
Microdeletion |
10 (30.3) |
3 (27.3) |
Duplication |
4 (12.1) |
7 (63.6) |
Microdel./Duplicat. |
ND |
1 (9.1) |
| |
|
|
|
|
|
|
|
*p<0.05; ND-not detected; Microdel./Duplicat.-Microdeletion/Duplication
table.jpg)
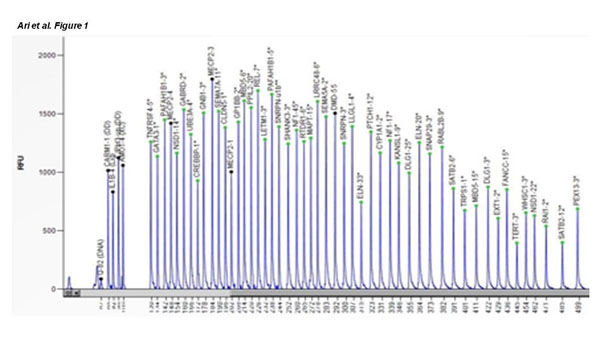
Figure 1. Shows the SALSA-MLPA probemix that designed for detection of microdeletions/duplications pik profiles in normal apperance for a female healthy individual as a control for the current study. All piks for target genes were in normal lenght and apperance
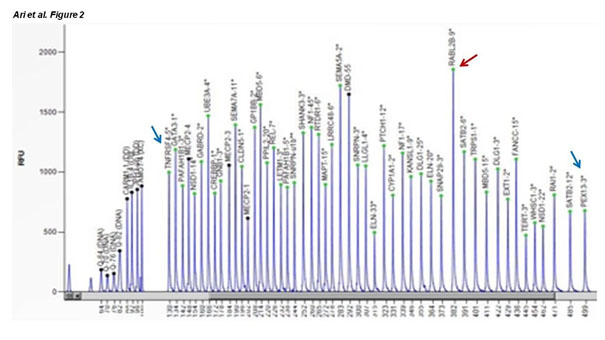
Figure 2. Red arrow shows the heterozygous duplicated and blue arrows show the heterozygous deleted genes in one of the current aborted material
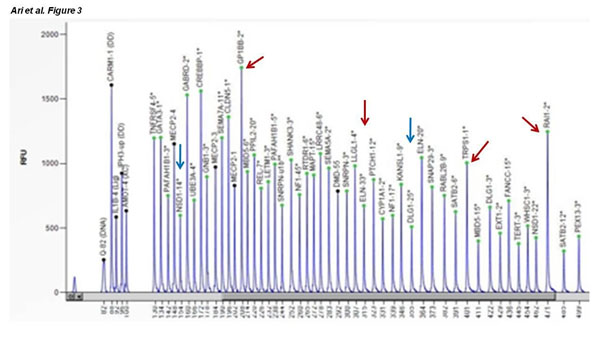
Figure 3. Red arrows show the more than two heterozygous duplicated and blue arrows show the heterozygous deleted genes in one of the current aborted material
When we compare the obtained results from two different tests (QF-PCR and MLPA aneyploidy test) we noted that 1 (1.5%) amnion sample were negative by QF-PCR analysis but positive by MLPA. In 98.51% the results were the same in both tests. In aborted material group, 2 aborted material samples (18.2%) were negative by QF-PCR but positive by MLPA analysis, so the results of these two tests was in agreement with 98.5% for the amnion samples and 81.8% for the aborted material samples. The rest of other samples were diagnosed as the same as QF-PCR technique.
Discussion
Molecular aneuploidy testing is a major benefit for pregnany women at increased risk of chromosomal abnormalities, leading to rapid reassurance for those with a euploid fetus and earlier decisions about further pregnancy management in case of abnormalities. Both QF-PCR and MLPA allow for detection of common aneuploidies within 24-48h and both methods have the inherent limitation of not being able to detect structural chromosome aberrations and all masaics. MLPA have several advantages compared to QF-PCR. In MLPA, 36 oligonucleotide probes are used to detect 36 loci in a single reaction whereas QF-PCR is only limited to approximately 12 loci or less. Moreover, increasing primers in multiplex PCR can cause problems due to primer-dimer interactions. Furthermore, MLPA uses non-polymorphic markers that is highly likely to be present in general population while the QF-PCR uses polymorphic short tandem repeat markers which show variable frequency in different populations and some of these markers can be non-informative if the patient is homozygous for that allele [7-15]. MLPA technique has some additional limitations. Bruno et al. have performed study on spontaneous abortion samples using FISH in conjunction with MLPA to detect polyploidy which can account for up 13% of abnormalities in spontaneous abortions [16]. The MLPA has been confirmed as a rapid, simple and reliable method for detection of chromosome 13, 18, 21, X and Y abnormalities in fetal tissue also in some of the previously studies in miscarriages [17]. The advantages of MLPA methos is now widely accepted, such as lower failre rates, lower cost and faster time for results, but this must be complemented by adequate limitations of the methodology, including the inability to characterize balanced structural rearrangements, especially if MLPA is to be performed alone [12- 20].
In our study, we estimated the significantly different distribution of chromosomal abnormalities between amnion and aborted samples groups (p < 0.0001) which could be additional prove for miscarriage due to cytogenetic abnormalities. More than 50-70% of first trimester spontaneous miscarriages are due to chromosomal abnormalities, and about 95% of there are due to autosomal aneuploidies [21, 22]. In our aborted fetal material we can concluded that it could be due to the chromosomal abnormalities. Our results show a high level of agreement between QF-PCR and MLPA technique. Two aborted material and one amnion samples that were negative for aneuploidy by QF-PCR test were positive by MLPA test. The rest of samples were diagnosed as the same as QF-PCR technique.
Conclusion
In conclusion, the results of the study confirmed the chromosomal abnormalities as a reason for abortion and showed that MLPA is comparable with that of QF-PCR for the detection of common aneuploidies and could be another rapid and reliable tool for prenatal diagnosis. Both MLPA kits can be used for the prenatal diagnosis successfully, but it is better to use it with combination of the other techniques or results need to be confirmed by other valid techniques such as QF-PCR, fully karyotyping, Sanger sequencing and/or microarray in the prenatal diagnosis.
Competing interests
All authors declare that they have no competing interests.
Acknowledgements
The authors thank the family members who agreed to participate in the presented results. Funding for this research was provided by Scientific Research Foundation Unit (BAP) of Canakkale Onsekiz Mart University, Canakkale-Turkey (Grant No: BAP - 2014/227).
Authors' contributions
E.A. did all experimental analysis and designed the clinical experiments; O.O. designed the experiments, analysed the data and critically review the manuscript; J.Dj. performed statistical analysis and wrote all parts of the manuscript, F.S. analysed the clinical data and designed the clinical experiments and give the final approval for manuscript.
All authors read and approved the final manuscript
References
- Ford HB, Schust DJ (2009) Recurrent pregnancy loss: etiology, diagnosis and therapy. Rev Obstet Gynecolx 2: 76-83. [Crossref]
- Gracia CR, Sammel MD, Chittams J, Hummel AC, Shaunik A, Barnhart KT (2005) Risk factors for spontaneous abortion in early symptomatic first-trimester pregnancies. Obstet Gynecol 106: 993-999. [Crossref]
- Garcia-Enguidanos A, Calle ME, Valero J, Luna S, Dominquez-Rojas V (2002) Risk factors in miscarriage: a review. Eur J Obstet Gynecol Reprod Biol 102: 111-119. [Crossref]
- Goddijn M, Leschot NJ (2000) Genetic aspects of miscarriage. Baillieres Best Pract Res Clin Obstet Gynaecol 14: 855-865. [Crossref]
- Lomax B, Tang S, Separovic E, Phillips D, Hillard E, et al. (2000) Comparative genomic hybridization in combination with flow cytometry improves results of cytogenetic analysis of spontaneous abortions. Am J Hum Genet 66: 1516-1521. [Crossref]
- Van der Berg MM, van Maarle MC, van Wely M, Goddijn M (2012) Genetics of early miscarriage. Biochim Biophys Acta 1822: 1951-1959. [Crossref]
- Badenas C, Rodriques-Revenga L, Morales C, Mediano C, Plaja A, et al. (2010) Assessment of QF-PCR as the first approach in prenatal diagnosis. J Mol Diagn 12: 828-834. [Crossref]
- Langlois S, Duncan A (2011) SOGC GENETICS COMMITTEE; CCMG PRENATAL DIAGNOSIS COMMITTEE. Use of a DNA method, QF-PCR, in the prenatal diagnosis of fetal aneuploidies. J Obstet Gynecol Can 33: 955-960. [Crossref]
- Massalska D, Bijok J, Zimowski JG, Jozwiak A, Jakiel G, Roszkowski T (2013) Multiple ligation-dependent probe amplification (MLPA)- new possibilities of prenatal diagnosis. Ginekol Pol 84: 461-464. [Crossref]
- Slater HR, Bruno DL, Ren H, Pertile M, Schouten JP, et al. (2003) Rapid, high throughput prenatal detection of aneuploidy using a novel quantitative method (MLPA). J Med Genet 40: 907-912. [Crossref]
- Saxena D, Agarwai M, Gupta D, Agarwai S, Das V Phadke SR (2016) Utility and limitations of multiplex ligation-dependent probe amplification technique in the detection of cytogenetic abnormalities in products of conception. J Postgrad Med 62: 239-241. [Crossref]
- Stuppia L, Antonucci I, Palka F, Gatta V (2012) Use of the MLPA assay in the molecular diagnosis of gene copy number alterations in human genetic diseases. Int J Mol Sci 13: 3245-3276. [Crossref]
- Hamidah HN, Munirah AR, Hafiza A, Farisah AR, Shuhaila A, et al. (2014) Prenatal diagnosis of aneuploidies in amniotic fluid by multiple ligation-dependent probe amplification (MLPA) analysis. Malays J Pathol 36: 163-168. [Crossref]
- Schouten J, Galjaard RJ (2008) MLPA for prenatal diagnosis of commonly occurring aneuploidies. Methods Mol Biol 444: 111-122. [Crossref]
- Van Opstal D, Boter M, de Jong D, van den Berg C, Bruggenwirth HT, et al. (2009) Rapid aneuploidy detection with multiplex ligation-dependent probe amplification: a prospective study of 4000 amnionic fluid samples. Eur J Genet 17: 112-121. [Crossref]
- Bruno DL, Burgess T, Ren H, Nouri S, Pertile MD, et al. (2006) High-throughput analysis of chromosome abnormality in spontaneous miscarriage using and MLPA subtelomere assay with an ancillary FISH test for polyploidy. Am J Med Genet A 140: 2786-2793. [Crossref]
- Carvalho B, Doria S, Ramalho S, Brandao O, Sousa M, et al. (2010) Aneuploidies detection in miscarriages and fetal deaths using multiplex ligation-dependent probe amplification: an alternative for speeding up results? Eur J Obstet Gynecol Reprod Biol 153: 151-155. [Crossref]
- Caramins MC, Saville T, Shakeshaft R, Mullan GL, Miller B, et al. (2011) A comparison of molecular and cytogenetic techniques for the diagnosis of pregnancy loss. Genet Med 13: 46-51. [Crossref]
- Kim JW, Lyu SW, Sung SR, Park JE, Cha DH, et al. (2015) Molecular analysis of miscarriage producst using multiplex ligation-dependent probe amplification (MLPA): alternative to conventional karyotype analysis. Arch Gynecol Obstet 291: 347-354. [Crossref]
- Kooper AJ, Faas BH, Feuth T, Creemers JW, Zondervan HH, et al. (2009) Detection of chromosome aneuploidy in chorionic villus samples by multiplex ligation-dependent probe amplification. J Mol Diagn 11: 17-24 [Crossref]
- Wolf CG, Horge EO (1995) Indications for examination of spontaneous abortion speciems: a reassessment. Am J Obstet Gynecol 173: 1364-1368.
- Rubio C, Pehlivan T, Rodrigo L, Simon C, Remohl J, Pellicer A (2005) Embryo aneuploidy screening for unexplained recurrent miscarriage: a minireview. Am J Reprod Immunol 53: 159-165. [Crossref]

table.jpg)


