Objectives: In this study we investigated the susceptibility profiles against macrolides and lincosamides of 85 C. striatum strains isolated at a clinical centre in Sousse (Tunisia).
Methods: The strains were identified by the routine biochemical assays and then confirmed by Vitek-Maldi-Tof-MS. MIC’s of erythromycin and clindamycin were determined using the microdilution method. The detection of erm(X), erm(B), msr(A), mph(A) and mef(A-E) resistance genes was performed by PCR. The strains were typed by PFGE using XbaI.
Results: Sixty-nine (81.17%) strains were resistant to erythromycin, 58 (67.44%) strains were resistant to clindamycin. There was a high correlation between the resistance to erythromycin and clindamycin and the presence of erm(X) gene in 85.50% and 89.65% respectively. The erm(B) gene was detected in 21(24.70%) strains whereas, no others genes were detected in our strains collection. By PFGE, the 85 strains belonged to 18 different clones.
Conclusion: erm(X) is implicated in macrolide resistance for almost all the Corynebacterium strains analyzed in our study. Other resistance genes like erm(B) must also be implicated in this resistance, although its presence seems to be unusual in previously reported studies.
Corynebacterium striatum, macrolide, Lincosamide, resistance, erm genes, PFGE
Corynebacterium striatum is an ubiquitous microorganism of the human skin and mucous membranes. However, it is a significant emerging opportunistic pathogen causing serious infections in immunocompromised and hospitalized patients and isolated as the etiological factor of various infections such as pneumonia vertebral osteomyelitis septicemia and endocarditis [1-4]. Different studies showed high resistance rates against several antimicrobials and many outbreaks of multidrug resistant C. striatum have been recently reported [5,6]. Macrolides are frequently used in infections that can be caused by these microorganisms and are an alternative to b-lactams and tetracycline. They act by binding to the 50S subunits of bacterial ribosomes and interfere with protein synthesis by inhibiting the elongation of peptide chains [7]. Macrolide resistance in C. striatum is of particular interest because it is clearly an acquired phenotype and is common in clinical isolates. In C. striatum, resistance to macrolides was previously shown to be mainly due to methylase enzymes that are codified by erm genes.
Other resistance mechanisms such as mutations in 23S rRNA domain V, modifications of proteins L4 and L22, efflux pump systems, and hydrolytic enzymes have been reported in different genera [8,9]. Several studies reported high levels of resistance to macrolides among Corynebacterium species [10,11,12]. However, in Tunisia, no data are available. Therefore, we investigated the susceptibility profiles against macrolides and lincosamides of 85 C. striatum strains isolated at a clinical centre in Sousse (Tunisia).
Bacterial strains
We tested 85 C. striatum strains isolated from different specimens of patients presented at the laboratory of microbiology at the tertiary Hospital F. Hached Sousse, Tunisia, between January 2007 and December 2012. In this study, one isolate per patient was used. Strains were identified using a phenotypic method with the API Coryne system (bioMerieux) according to the manufacturer’s instructions and by comparison with the API WEB database. Strains were also identified using a MALDI-TOF/ VITEK MS mass spectrometry system (bioMerieux). Obtained spectra were analyzed with the VITEK MS database
Antimicrobial susceptibility testing
In the screening tests for all investigated strains, inducible type of MLSB resistance was tested. Antibiotic discs (Biorad) of erythromycin (E - 15 mg) and clindamycin (Cln- 2 mg) were placed 15–20mm from each other on the on cation-adjusted Mueller-Hinton agar with 5% sheep blood and inoculums of 1.0 McFarland. The plates were incubated for 48 hours at 35°C. The appearance of a flattening zone around the clindamycin confirmed the presence of inducible resistance. In the case that approved breakpoints for Corynebacterium spp. were absent, criteria for MLSB antibiotics for Staphylococcus spp were used. The susceptibility to erythromycin and clindamycin was also investigated using microdilution method in cation adjusted Muller-Hinton broth in accordance with guidelines of the Clinical and Laboratory Standards Institute (CLSI). The minimum inhibitory concentration (MIC) was estimated according to the CLSI guidelines for Corynebacterium spp. In the absence of approved breakpoints for Corynebacterium spp, breakpoints reported for Staphylococcus spp were used. C. striatum (ATCC 6940), C. jeikeium (ATCC 43734), E. coli (ATCC 25922) and S. pneumoniae (ATCC 49619) and S. aureus (ATCC 29213) strains were used as controls. The strains were grown on blood agar plates during 24–48 hours at 37°C in normal atmosphere and checked for purity.
Nucleic acid extraction and PCR
Few colonies were suspended in 200 ml of Instagene matrix (Biorad). The bacterial suspension was incubated during 15 minutes at 56°C followed by 8 minutes at 100°C and subsequently centrifuged at 12,000 rpm for 3 minutes. Supernatant was used for PCR studies. PCR was performed using commercial KAPA2G Fast HotStart Ready Mix (2X). The primers used for detection of the erm(X) gene were Cerm 1 (5’-GAC ACG GCC GTC ACG AGC AT-3’) and Cerm 2 (5’-GGC GGC GAG CGA CTT CC- 3’); for the erm(A) gene: Erm A-1 (5’-TCT AAA AAG CAT GTA AAA GAA-3’) and Erm A-2 (5’-CTT CGA TAG TTT ATT AAT AGT-3’); for the erm(B) gene: Erm B-1 (5’-GAA AAG GTA CTC AAC CAA ATA-3’) and Erm B-2 (5’-AGT AAC GGT ACT TAA ATT GTT TAC-3’); and for the mef(A-E) gene: Mef A-E-1 (5’-AGT ATC ATT ATT CAC TAG TGC-3’) and Mef A-E-2 (5’-TTC TTC TGG TAC TAA AAG TGG-3’) and for the mph(A) gene: MphA-1(5’-AAC TGT ACG CAC TTG C-3’) and MphA-2 (5’-GGT ACT CTT CGT TAC C-3’); and for the msrA gene: MsrA-1 (5’ GCA CTT ATT GGG GGT AAT GG3’) and MsrA-2 ( 5’GTC TAT AAG TGC TCT ATC GTG 3’). The erm(X) gene amplification was performed according to the protocol described by Rosato, et al. [10]. For all other genes we applied PCR conditions previously reported for Streptococcus sp [13]. The amplification products were detected by electrophoresis in 2% agarose. The size of the amplicons was estimated using commercial molecular weight ladder (BioRad Laboratories, Hercules, CA).
Pulsed-field gel electrophoresis (PFGE)
For further characterization of C. striatum isolates, we used the PFGE macro-restriction analysis with XbaI as previously described [5]. Macro-restriction fragments were separated on 1% (wt/vol) ultrapure agarose gels at 6V/Cm, for 18h at 14°C with pulse times of 0.1-5 s on CHEF-DR® III. Lambda DNA concatemers were used as molecular size markers. Similarities among macro-restriction pattern were identified according to established criteria.
A total of 85 C. striatum strains were isolated from different specimens (Table 1). The API Coryne often provided ambiguous results and could not differentiate between C. striatum and other related species such as C. amycolatum, C. aurimucosum, C. minutissimum and C. simulans. However, in the MALDI-TOF-MS method the strains were identified with cut off ≥99.99. Screening test for sensitivity to erythromycin and clindamycin was performed using the disc diffusion method. MIC determinations for erythromycin and clindamycin with microdilution method confirmed the results of the disc diffusion method.
Table 1. Isolated strains of C. striatum (n = 85) from various infection sites
Source |
Number of species |
Percentage % |
Wound swab |
20 |
23.53% |
blood |
3 |
3.53% |
Eye conjunctival swab |
7 |
8.23% |
Ear swab |
15 |
17.65% |
Sputum |
5 |
5.89% |
Urine |
9 |
10.58% |
Vaginal swab |
8 |
9.41% |
Tracheostomy secretion |
6 |
7.06% |
Catheter injection site |
4 |
4.70% |
Sperm |
3 |
3.53% |
Medical devices |
5 |
5.89% |
Resistance to examined antibiotics was observed in approximately 80% of the strains, ten of which appeared to be resistant only to one antibiotic: seven were resistant to clindamycin, while three were resistant to erythromycin. Fifty-three strains showed resistance to both antibiotics (Table 2). The shape of the growth inhibition zones indicated the presence of a constitutive MLSB resistance type.
Table 2. Susceptibility of 85 C. striatum strains studied to MLS antibiotics
|
Range (µg/ml) |
Susceptibility, number of strains (%) |
Antibiotics |
|
S |
I |
R |
Erythromycin |
0.06-64 |
14 |
2 |
69 |
Clindamycin |
0.06-64 |
7 |
20 |
58 |
S: Susceptible, R: Resistant, I: Intermediate
The bacterial DNA of all 85 strains was amplified with primers for resistance genes to macrolide and lincosamide antibiotics. In 62 out of 85, the erm(X) gene was present. The amplification product of erm(B) gene was detected in 21 C. striatum strains, and in 12 strains both erm(X) and erm(B) amplicons were detected. There was a high correlation between the resistance to erythromycin and clindamycin and the presence of erm(X) gene in 85.50% and 89.65% respectively (Figure 1). Whereas, the erm(B) gene was detected in 26.68% and 20.68% of the strains resistant to erythromycin and clindamycin respectively (Figure 1). No amplification products of other tested genes were detected in any of the studied strains (Table 3). Although they showed resistance to macrolide and clindamycin, no amplification products were detected in 3 strains.
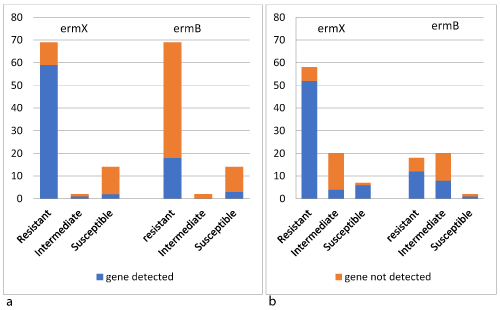
Figure 1. Distribution of erm(X) and erm(B) genes among C. striatum strains according to the antimicrobial agents tested: (a) Erythromycin; (b) Clindamycin
Table 3. Antibiotic susceptibility patterns of C. striatum resistant isolates and their genotypes
|
|
Genes |
E and Cln susceptibility phenotype (MIC) |
N° of strains (%) |
ermX |
ermB |
mefA-E, mphA, msrA |
ER ClnR |
53 (62.35%) |
50 |
12 |
- |
ER ClnS |
7 (8.23%) |
6 |
1 |
- |
ES ClnR |
3 (3.52%) |
1 |
0 |
- |
EI ClnR |
2 (2.35%) |
1 |
0 |
- |
ER ClnI |
9 (10.58) |
3 |
5 |
- |
ES ClnI |
11(12.94%) |
1 |
3 |
- |
Total |
85 (100%) |
62 |
21 |
0 |
ER ClnR, erythromycin and clindamycin-resistant strains.
ER ClnS, erythromycin-resistant and clindamycin-susceptible strains.
ES ClnR, erythromycin-susceptible and clindamycin-resistant strains.
ER ClnI, erythromycin-resistant and clindamycin- intermediate susceptibility strains.
EI ClnR, erythromycin-intermediate susceptibility and clindamycin-resistant strains
ES ClnI, erythromycin-susceptible and clindamycin-intermediate susceptibility strains
PFGE analysis indicated that the 85 C. striatum isolates represented 18 distinct strain types (data not shown), with the forty ERY-susceptible strains being distinct from the ERY resistant strain types. Ten strains resistant to erythromycin, isolated from the same ward in the same period, belonged to the same clone.
The genus Corynebacterium includes a high number of species that are usually isolated from human skin as saprophytes. However, these microorganisms have also been reported as infectious agents in a broad group of patients and have showed broad-spectrum resistance.
In this study, including 85 strains of C. striatum derived from a variety of infections in humans, the use of conventional method based on the morphological and biochemical traits (Api Coryne strips) proved to be insufficient and only the application of the MALDI-TOF/VITEK method allowed the correct identification of the strains.
The C. striatum strains studied showed high resistance to macrolides of (81.17%), a fact that confirms the previously reported high prevalence of resistance among Corynebacterium species [10,11,14,15]. Clindamycin showed low activity against C. striatum strains (67.44%). The activity of erythromycin and clindamycin was almost identical. Among Corynebacterium species, the most frequently described macrolide resistance mechanism is the presence of methylase enzymes codified by erm(X) class genes.
The results obtained in our study confirm the high prevalence of the erm(X) genes and the correlation between the presence of this amplicon and the MLSB phenotype was higher than 80%. Three strains showed an MLSB resistance phenotype despite being negative for all genes tested. This must be due to the presence of the other resistance mechanisms. In this sense, mutations in 23S rRNA domain V and alterations of protein L4 and L22 have originated an MLSB resistance phenotype [16-19].
The amplification products of the erm(B) gene were detected in 21 (24.70%) strains. The presence of this gene has already been described in group A Corynebacterium strains [20-22]. In this study, the prevalence of this gene was higher than previously reported by Ortiz-Pérez, et al. [15] and Szemraj et al. [11].
Besides, there were 12 strains that carried the erm(X) and erm(B) genes simultaneously, a phenomenon previously detected in C. urealyticum and in Staphylococcus where the presence of several erm genes is very common [23].
In our study, no amplification products of the mef(A-E) gene were detected, although this gene has been previously reported in the genus Corynebacterium [22]. Other efflux pump genes such as msr have also been reported in this genus [24], but they were not detected in our report. PFGE analysis showed that the strains tested were genetically unrelated. The 10 resistant strains belonging to the same clone and isolated from the same ward in the same period suggested a clonal outbreak nosocomial infection.
According to our data erm(X) is implicated in macrolide resistance for almost all the C. striatum strains analyzed in our study. Other resistance genes like erm(B) must also be implicated in this resistance, although its presence seems to be unusual in previously reported studies.
None declared.
Not required.
- Tarr PE, Stock F, Cooke RH, Fedorko DP, Lucey DR (2003) Multidrug-resistant corynebacterium striatum pneumonia in a heart transplant recipient. Transpl Infect Dis 5: 53-58. [Crossref]
- Ferndez-Ayala M, Nan N, Faris MC (2001) Vertebral osteomyelitis due to corynebacterium striatum. Am J Med 111: 160-167. [Crossref]
- Martin MC, Meln O, Celada MM, Alvarez J, Mandez FJ (2003) Septicaemia due to corynebacterium striatum: Molecular confirmation of entry via the skin. J Med Microbiol 52: 599-602. [Crossref]
- de Arriba JJ, Blanch JJ, Mateos F, Martínez-Alfaro E, Solera J (2002) Corynebacterium striatum first reported case of prosthetic valve endocarditis. J Infect 44: 193. [crossref]
- Campanile F, Carretto E, Barbarini D, Grigis A, Falcone M, et al. (2009) Clonal multidrug-resistant Corynebacterium striatum strains, Italy. Emerg Infect Dis 15: 75-78.
- Baio PV, Mota HF, Freitas AD, Gomes DL, Ramos JN, et al. (2013) Clonal multidrug-resistant Corynebacterium striatum within a nosocomial environment, Rio de Janeiro, Brazil. Mem Inst Oswaldo Cruz 108: 23-29. [Crossref]
- Weisblum B (1995) Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother 39: 577-585.
- Nakajima Y (1999) Mechanisms of bacterial resistance to macrolide antibiotics. J Infect Chemother 5: 61-74. [Crossref]
- Tait-kamradt A, Davies TA, Cronan M, Jacobs MR, Appelbaum PC, et al. (2000) Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob Agents Chemother 44: 2118-2125.
- Rosato AE, Lee BS, Nash KA (2001) Inducible, macrolide resistance in corynebacterium jeikeium. Antimicrob Agents Chemother 45: 1982-1989. [Crossref]
- Szemraj M, Kwaszewska A, Pawlak R, Szewczyk EM (2014) Macrolide, lincosamide, and streptogramin B resistance in lipophilic corynebacteria inhabiting healthy human skin. Microb Drug Resist 20: 404-409. [Crossref]
- Olender A (2013) Antibiotic resistance and detection of the most common mechanism of resistance (MLSB) of opportunistic corynebacterium. Chemotherapy 59: 294-306. [Crossref]
- Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L (1996) Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother 40: 2562-2266.
- Soriano F, Zapardiel J, Nieto E (1995) Antimicrobial susceptibilities of corynebacterium species and other nonspore- forming gram-positive bacilli to 18 antimicrobial agents. Antimicrob Agents Chemother 39: 208-214.
- Ortiz-Pez A, Martin-de-Hijas NZ, Esteban J, Ferndez-Natal MI, Garca-Ca JI, et al. (2010) High frequency of macrolide resistance mechanisms in clinical isolates of corynebacterium species. Microbial Drug Resist 16: 273-277. [Crossref]
- Weisblum B (1995) Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother 39: 577-585.
- Leclercq R, P Courvalin (1991) Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob Agents Chemother 35: 1267-1272. [Crossref]
- Leclercq R, P Courvalin (1993) Mechanisms of resistance to macrolides and functionally related antibiotics. In Bryskier AJ, Butzler JP, Neu HC, Tulkens PM (Ed.), Macrolides chemistry, pharmacology and clinical uses. Arnette, Blackwell, Paris.
- Wei L, Zheng Z (2013) Implication of ermCX gene of corynebacterium striatum in macrolide resistance in Beijing, China. Bangladesh J Pharmacol 8: 54-57
- Yague G, Mora B, Martnez-Toldos MC, Rodriguez T, Valero PL, et al. (2005) Implication or ermX genes in macrolide- and telithromycin-resistance in Corynebacterium jeikeium and corynebacterium amycolatum. Rev Esp Quimioter 18: 2362-242. [Crossref]
- Luna VA, Coates P, Eady EA, Cove JH, Nguyen TT, et al. (1999) A variety of gram-positive bacteria carry mobile mef genes. J Antimicrob Chemother 44: 19-25.
- Aktas Z, Aridogan A, Kayacan CB, Aydin D (2007) Resistance to macrolide, lincosamide and streptogramin antibiotics in staphylococci isolated in Istanbul, Turkey. J Microbiol 45: 286-290.
- Ojo KK, Striplin MJ, Ulep CC, Close NS, Zittle J, et al. (2006) Staphylococcus efflux msr(A) gene characterized in streptococcus, enterococcus, corynebacterium, and pseudomonas isolates. Antimicrob Agents Chemother 50: 1089-1091. [Crossref]
- Roberts MC, Sutcliffe J, Courvalin P, Jensen LB, Rood J, et al. (1999) Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob Agents Chemother 43: 2823-2830.

