Abstract
Thyroid cancer is the most common type of endocrine-related cancer globally. Although considered a relatively rare disease, in last decades, the number of new cases of thyroid cancer has grown faster than other malignancies. Although 70-80% of all registered cases comprise the papillary and follicular thyroid cancer, which show a survival rate in five years upper to 98%, near 30% of patients present a history of recurrent disease and 10%, dediferiation to most agreessive disease forms: poor differentiated and anaplastic thyroid cancer (the most lethal cancer known). Thus, to understand the molecular mechanisms that regulate the epithelial-mesenchymal transition (EMT) is mandatory to identify disseminating cancer cells before the metastasis formation, as well as to propose most effective therapies for patients with recurrent and/or metastatic disease history. In this context, LIMD2 emergers as a useful biotechnological target, since the gene (LIMD2) is differentially expressed in primary tumors and metastasis from PTC. For this reason, to study the biology of LIMD2 in EMT can bring new opportunities in both diagnostic and therapeutic field.
Keywords
thyroid cancer, metastasis, epithelial-mesenchymal transition, LIM domain, LIMD2
Thyroid cancer
Thyroid cancer comprises the most common type of endocrine-related cancer [1,2]. Although considered a rare disease, in last decades the rate of disease incidence has been increasing alarmingly, specially in Italy [3], France, Israel, Australia [4], South Korea [5,6] and United States, countries where the incidence increased more than three folds [5,7-10]. According to the National Cancer Institute (NCI), an expected 53,990 new cases of thyroid cancer in 2018 in United States (https://seer.cancer.gov/statfacts/html/thyro.html). The average annual percentage thyroid cancer is increasing dramatically in both men (5,4%) and women (6,5%). The 2020 and 2030 incidence projection shows that the incidence of thyroid cancer will surpass that of colorectal cancer by 2030 and will be the leading cancer site in 2030 for women [11].
In Brazil, the estimated number of new cases of female thyroid cancer women was 8.040 per 100,000 women per year, being the fifth most prevalent cancer in females (http://www.inca.gov.br).
Despite this increasing incidence, the mortality rate by thyroid cancer remains stable. However, current studies showed an increase in both thyroid cancer-related incidence and mortality in USA among 1973-2013 [12-14]. By combining these data, the authors evaluated the incidence-based thyroid cancer mortality rates according tumor size, stage, histology at diagnosis. The authors pointed out that the incidence and mortality also increased for advanced stage tumors, suggesting a real increase incidence of the disease and not only a consequence of the overdiagnosis [5,15].
Most primary thyroid cancer originate from thyroid follicular cells [16], which encompass the differentiated (papillary and follicular thyroid carcinoma), the poorly-differentiated thyroid carcinoma (PDTC) and the undifferentiated or anaplastic thyroid carcinoma (ATC) [16,17]. Papillary thyroid carcinoma (PTC) comprises nearly most (80-90%) of all thyroid cancer cases, followed by follicular thyroid carcinoma (FTC) (5-10%) [9]. Medullar thyroid cancer (MTC) is a rare disease (2%) that arises from parafollicular cells [16,17].
As most types of cancers, thyroid cancer is associated with the effect of multiple genes, often coupled with lifestyle and environmental factors. Risk factors for PTC includes familial history of thyroid disease or thyroid cancer, inherited conditions, radiation exposure and gender, and for FTC a low-iodine diet and familial conditions [18,19-21].
Untill the 1990’s decade, it was estimated that 25% of thyroid carcinomas were related to mutations [22]. As most thyroid carcinomas have an indolent clinical behavior, it is expected that the tumors have a stable genome and low mutational burden. In 2014, through the The Cancer Genome Atlas (TCGA), the International Cancer Genome Consortium that performed genomic analysis of near 500 PTC, the genetic landscape of most PTC was elucidated and this percentage increased to 90% [23,24]. Although multiple known genes were associated with pathogenesis of PTC, the authors confirmed that thyroid cancer is a genetically quiet disease with a relatively low number of mutations in each cancer. Recurrent point mutation or gene fusion in know genes that activates the MAPK (i.e., BRAF, RAS, RET/PTC and NTRK) and PI3K/AKT/mTOR (i.e., signaling pathways PAX8/PPARG, PTEN) [25]. The authors also described recurrent mutations in novel genes (EIF1AX) or new fusion from old oncogenes (RET, BRAF, NRTK1/3, ALK, and FGFR2), as well as copy number alteration of 22q [26,17,26-31]. The TCGA data also confirmed the presence of somatic point mutations in the promoter region of the telomerase reverse transcriptase gene (TERT) in 9% of PTC. Tumor with BRAF V600E or RAS and TERT mutations are associated with high risk of recurrence, loss of thyroid differentiation with consequent loss of radioiodine avidity, and higher risk of mortality.
As the TCGA data also showed that the BRAF and RAS-mutated PTC showed a mutually exclusive expression pattern, the authors developed a BRAFV600E - RAS score, termed BRS. Using BRS grade, all other samples were classified according biological features of RAS (Ras-like) or BRAFV600E (BRAFV600E –Like). Additionally, using the expression of 16 thyroid- related genes, a thyroid differentiation score (TDS) was established to measure thyroid-specific differentiation. Fusions of the PAX8 and PPARG genes and RAS mutations are common in FTC.
Current management of thyroid cancer involves determination of risk of recurrence, in order to bet treatment plan. Up to now, there is no reliable clinical, pathological or molecular markers that can distinguish which patients with thyroid carcinoma is candidate for active surveillance or will progress and develop clinical significant disease and, therefore, should undergo surgery (lobectomy vs total total thyroidectomy), followed by radioactive iodine therapy and suppressive therapy.
Additionally, although the majority of the cases have a satisfactory responds to conventional therapy, a subset of DTC, PDTC and ATC are refractory to standard treatment [17]. Moreover, about 10% of DTC progress to PDTC or ATC [32], which exhibits a poor prognostic due to the metastatic potential .
In order to develop new therapeutic strategies for RAI-refractory thyroid carcinomas, pre-clinical and clinical studies have been conducted using tyrosine kinase inhibitors (TKIs) acting at the level of signaling transduction pathways (i.e., RAS, BRAF/MEK/ERK, RET/PTC and VEGF receptors). Some of these TKIs may restore iodine uptake and clinical response to RAI (please see recent review),
Despite advances using TKIs, which are effective against specific genetic mutations, metastasis still persists as a barrier to successful therapy, as secondary mutation in the target gene and activation of alternative pathways may occur. Beyond these mechanisms of drug resistance, most of genetic associated to metastatic process are unknown and phenotypic transformation (epithelial-mesenchymal transition) may represent additional mechanism of drug resistance.
In fact, several studies have suggested that both cancer metastasis and drug cancer resistance are related to a complex pathological process known as the epithelial-mesenchymal transition (EMT), This process involves genetics, biochemical and morphological deregulations that lead to downregulation of epithelial markers and an upregulation of mesenchymal markers as a consequence of aberrant activation of signaling molecules and transcription factors [33]. These changes lead to an acquisition of a hybrid phenotype (epithelial-mesenchymal), which exhibits loss of cell polarity and depolymerization in actine fibers, resulting in a fibroblastoid morphology acquisition.
Genetic reprograming verified during the EMT not only lead to cancer cell dissemination, but also contributes with the cancer-stem cell (CSC) phenotype acquisition as a consequence of the energetic metabolism deregulations [34].
Thus, to identify genes differentially expressed or mutated during metastatic process is crucial for new treatment strategies. Additionally, it may help also the diagnostic of circulating cancer cells (liquid biopsy).
LIMD2: A biotechnological target for diagnostic and immunotherapy
Among the several genes deregulated on thyroid cancer, LIMD2 occupies a prominent position once it is differentially expressed during metastatic process and regulates cell motility and tumor progression in different tumor subtypes, including thyroid [35-37].
Gene LIMD2 (locus 17q23.3) codifies a small adapter protein (LIMD2 - LIM domain containing 2) able to regulate different biological process, including cell motility [36]. Seven splicing variants of LIMD2 were described (Table 1).
Table 1. LIMD2 splicing variants. Amino acid sequence of LIMD2 variants showing the antigen sequence (GFGRKQHKELWAHKEVDPGTKT) in bold. Source: Protein Atlas.
Variant |
UniProt |
Size |
Weight |
Sequence |
LIMD2-001 |
Q9BT23 |
127 aa |
14,070 kDa |
MFQAAGAAQATPSHDAKGGGSSTVQRSKSFSLRAQVKETCAACQKTVYPMERLVADKLIFHNSCFCCKHCHTKLSLGSYAALHGEFYCKPHFQQLFKSKGNYDEGFGRKQHKELWAHKEVDPGTKTA |
LIMD2-002 |
Q9BT23 |
127 aa |
14,070 kDa |
MFQAAGAAQATPSHDAKGGGSSTVQRSKSFSLRAQVKETCAACQKTVYPMERLVADKLIFHNSCFCCKHCHTKLSLGSYAALHGEFYCKPHFQQLFKSKGNYDEGFGRKQHKELWAHKEVDPGTKTA |
LIMD2-003 |
J3QL69 |
78 aa |
9,028 kDa |
MERLVADKLIFHNSCFCCKHCHTKLSLGSYAALHGEFYCKPHFQQLFKSKGNYDEGFGRKQHKELWAHKEVDPGTKTA |
LIMD2-006 |
J3QL69 |
78 aa |
9,028 kDa |
MERLVADKLIFHNSCFCCKHCHTKLSLGSYAALHGEFYCKPHFQQLFKSKGNYDEGFGRKQHKELWAHKEVDPGTKTA |
LIMD2-007 |
J3QQM5 |
12,013 kDa |
MFQAAGAAQATPSHDAKGGGSSTVQRSKSFSLRAQVKETCAACQKTVYPMERLVADKLIFHNSCFCCKHCHTKLSLGSYAALHGEFYCKPHFQQLFKSKGNYDEGFGRK |
LIMD2-010 |
J3KRR0 |
87 aa |
9,518 kDa |
MFQAAGAAQATPSHDAKGGGSSTVQRSKHCHTKLSLGSYAALHGEFYCKPHFQQLFKSKGNYDEGFGRKQHKELWAHKEVDPGTKTA |
LIMD2-011 |
Q9BT23 |
127 aa |
14,070 kDa |
MFQAAGAAQATPSHDAKGGGSSTVQRSKSFSLRAQVKETCAACQKTVYPMERLVADKLIFHNSCFCCKHCHTKLSLGSYAALHGEFYCKPHFQQLFKSKGNYDEGFGRKQHKELWAHKEVDPGTKTA |
Due to its capability to bind proteins unable to bind themselves (adapter) [35], LIMD2 is a useful candidate as a target for novel therapeutics against thyroid cancer.
Although the three-dimensional structure of human LIMD2 protein had been solved by Faooji, et al. using nuclear magnetic resonance (NMR, available in https://www.ebi.ac.uk/pdbe/entry/pdb/2LZU), there is little information about its biological function.
LIMD2 protein has 127 amino-acids, a molecular weight from 9.028 to 14.040 kDa, according to the splicing variant (Table 1) and belongs to the LIM domain superclass [36,38,39].
The LIM domain was discovery about 30 years ago as a evolutionarely conserved zinc-finger motif, found in plants, fungi, amoeba and animals, including humans [38,40]. This domain comprises a region of 50-65 amino-acids, characterized by the presence of two zinc-fingers rich in cysteine-histidine residues, separeted by an hidrophobic binding (CX2CX17HX2C)-X2-(CX2CX17CX2C/H/D), as showed in Figure 1 [38,41].
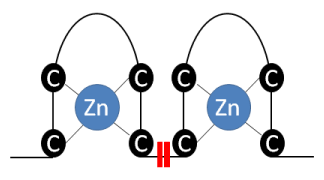
Figure 1. Representative scheme of zinc-finger, comprised by two regions rich in cysteine residues able to bind to zinc ions. This structure is connected by two hydrophobic amino acids.
LIM domain of LIMD2 protein is contained in the A or α chain, located among the residues 33-104 of the polipeptide, with 72 amino-acids lenght and a molecular weight of 8.32 kDa (Figure 1B) [38]. LIMD2 chain A exhibits both hydrophilic and hydrophobic regions, as showed by in silico analysis (Figure 2), which are related to the adapter function. Moreover, the chain A contains the most antigenic regions, with is comprised by the amino acid sequence (GFGRKQHQELWAHKEVDPGTK), showed in Table 1 and Figure 3.
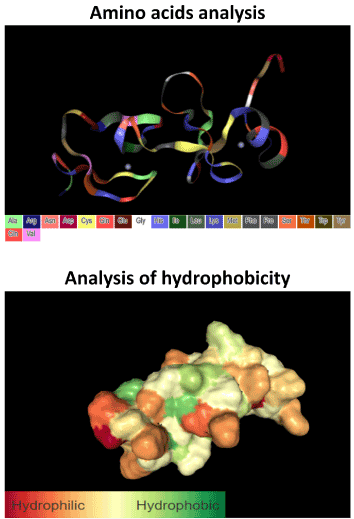
Figure 2. In silico analysis showing the amino acid position of LIMD2 tertiary structure and its hydrophobicity. LIMD2 exhibits both hydrophilic and hydrophobic regions, allowing its interaction with several proteins. Analysis performed through the Protein Data Bank tool.
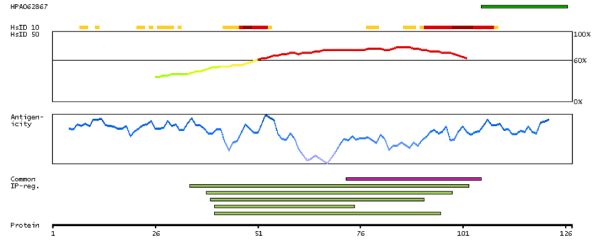
Figure 3. Analysis of LIMD2 antigenicity based on the splicing variant LIMD2-001 showing maximum sequence identity to other proteins from other human genes based on a 10 (HsID10) and 50 (HsID50) amino acid sliding window, the propensity to generate imune response based on nine amino acid sliding window (antigenicity) and interprotein regions. Data based on The Human Protein Atlas.
Proteins member of LIM family are very promiscuous. For this reason, is not surprising that LIMD2 protein be able to regulate different cellular functions, such as focal adhesion, adherents junction, cell proliferation, cyto-architeture and motility [38]. For this reason, LIM proteins, including LIMD2 is commonly upregulated in different malignancies, as showed in figure 4. Moreover, due its capability to deregulate cell adhesion complex and induces the motility, is not surprising that LIMD2 can be related to epitelial-mesenchymal transition (EMT) and, therefore, cancer metastasis [42].
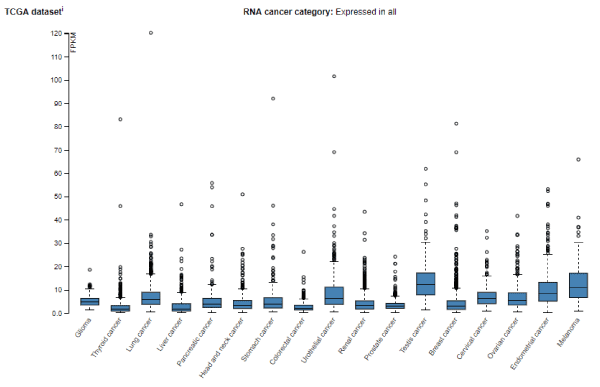
Figure 4. LIMD2 RNA expression in different human malignancies. Data obtained from The Human Protein Atlas.
Based on a phylogenetic study, Koch, et al. [38] analyzed 623 LIM domains from 265 proteins, clustering them according to patters of motif signatures in 14 superclasses: ABLIM, CRP, ENIGMA, EPLIN, LASP, LIMK, LHX, LMO, LMO7, MICAL, PXN, PINCH, TES and ZYN. Among these superclasses, EPLIN stands out to regulate cell motility though the repolymerization of actin fibers, affecting the cadherin-catenin complex integrity [38]. Due to the LIMD2 capability to interacts with cytoskeleton, promoting the actin polymerization disruption, the protein was classified as a member of EPLIN superclass [38].
The disruption of this complex is a hallmark of EMT, being also associated to the cancer-steam cell phenotype acquisition.
Based on this, studying paired paraffin-embedded tissue samples from primary PTC and metastasis, we verified a differential expression of LIMD2 in lymph node metastasis, in relation to primary PTC and normal thyroid, that did not show LIMD2 immunodetection [35,36]. We also verified that metastasis of PTCs with BRAF V600E, the most prevalent mutation verified in PTCs, show a LIMD2 overexpression when compared with metastasis from PTC with non mutated BRAF [35].
Considering that the cell cultures are useful models to study the EMT, we investigated the LIMD2 expression levels in cell lines derived from tumor thyroid cell line from PTC displaying BRAF V600E mutation (B-CPAP) and genetic fusion RET/PTC (TPC1), primary follicular thyroid carcinoma (FTC133) and lymph node metastasis related to this histological tumor type (FTC236) and, normal human thyroid (Nthy-ori 3-1) cell line. Results of these analyses showed the LIMD2 is exclusively expressed in B-CPAP and TPC1 cell lines [43], reinforcing that the LIMD2 expression is strictly related to PTC. Similar data were observed by Peng, et al. [37], that reported the protein expression in TPC1 cells.
Moreover, it was verified that the LIMD2 knockdown repress the migratory and invasive capability, as well as inhibits the colony formation in soft-agar assay [37]. By the opposite, the LIMD2 ectopic expression in urinary bladder cells led an increase of cell migration and invasiveness [37]. These data were corroborate by Ferguson, et al. [44], that showed that LIMD2 RNA expression was 4.7 fold higher in lymph nodes metastases compared to primary tumors from urinary bladder. Altogether, these data suggest that LIMD2 has a central role in metastatic process of thyroid and urinary bladder cancer.
Conclusion
Although the DTC has a farorable prognostic, about 30% of patients present historic of recurrent disease [5,6,10,15,45] and 10%, dedifferentiation for most aggressive forms of the disease (PDTC and ATC) [32]. Moreover, thyroid cancer can spread to lymph nodes and distant organs [15]. Thus, to study the biological mechanisms that regulate de thyroid cancer dissemination are mandatory to identify novel genes differentially expressed between primary tumors and metastasis. In this sense, LIMD2 emerges as a useful biotechnological target for both diagnosis and therapy, since the protein is differentially expressed in metastasis of PTC, histological type that comprise 90% of all thyroid cancers. Moreover, the gene expression is correlated with the mutation BRAF V600E, which represents the most common mutation verified in this malignancy. Thus, the studies based on LIMD2 open new frontiers in research of thyroid cancer metastasis.
Acknowledgement
The authors acknowledge the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, grant number 2014/06570-6 and 2017/14948-7) and Sociedade Brasileira de Endrocrinologia e Metabologia (SBEM, Research Grant 2018). JMC is a recipient of a scholarship of Research Productivity from CNPq.
References
- Wirth L, Fojo T (2015) In Manual de Oncologia de Harrison. Chabner B, Longo D (Eds.) pp. 813-823.
- Xing M (2005) BRAF mutation in thyroid cancer. Endocr Relat Cancer 12: 245-262. [Crossref]
- Dal Maso L, Panato C, Franceschi S, Serraino D, Buzzoni C, et al. (2018) The impact of overdiagnosis on thyroid cancer epidemic in Italy,1998-2012. Eur J Cancer 94: 6-15. [Crossref]
- Australian Institute of Health and Welfare1 (2018) Cancer in Australia: Actual incidence data from 1982 to 2013 and mortality data from 1982 to 2014 with projections to 2017. Asia Pac J Clin Oncol 14: 5-15. [Crossref]
- Shi L, DeSantis C, Jemal A, Chen A (2017) Changes in thyroid cancer incidence, post-2009 American Thyroid Association guidelines. Laryngoscope 10: 2437-2441
- Brito JP1, Hay ID2 (2017) Thyroid cancer: Overdiagnosis of papillary carcinoma - who benefits? Nat Rev Endocrinol 13: 131-132. [Crossref]
- Hahn L, Kunder C, Chen M, Orloff L, Desser T (2017) Indolent thyroid cancer: knowns and unknowns. Cancers Head Neck 2: 1.
- Zagzag J, Kenigsberg A, Patel KN, Heller KS, Ogilvie JB (2017) Thyroid cancer is more likely to be detected incidentally on imaging in private hospital patients. J Surg Res 215: 239–244.
- Osorio M, Moubayaed S, Su H, Urken M (2017) Adult height and head and neck cancer: A pooled analysis within the INHANCE Consortium. Head Neck 36: 1391.
- Poller D, Johnson S (2017) Recent developments in the pathology of thyroid cancer. Clin Oncol pp. 1-5.
- Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, et al. (2014) Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 74: 2913-2921.
- Davies L, Morris L, Hankey B (2017) Increases in Thyroid Cancer Incidence and Mortality. JAMA 318: 389-390. [Crossref]
- Lim H, Devesa SS, Sosa JA, Check D, Kitahara CM (2017) Trends in Thyroid Cancer Incidence and Mortality in the United States, 1974-2013. JAMA 317: 1338-1348. [Crossref]
- Kitahara CM, Devesa SS, Sosa JA (2017) Increases in Thyroid Cancer Incidence and Mortality-Reply. JAMA 318: 390-391. [Crossref]
- Davies L, Welch HG (2006) Increasing incidence of thyroid cancer in the United States, 1973-2002. JAMA 295: 2164-2167. [Crossref]
- Papewalis C, Ehlers M, Schott M (2010) Advances in cellular therapy for the treatment of thyroid cancer. J Oncol 2010: 179491. [Crossref]
- Morrison JA, Pike LA, Lund G, Zhou Q, Kessler BE, et al. (2015) Characterization of thyroid cancer cell lines in murine orthotopic and intracardiac metastasis models. Horm Cancer 6: 87-99. [Crossref]
- SCHLUMBERGER M, PACINI F (1999) In Thyroid Tumors. Schlumberger M, Pacini F (Eds.) Nucléon.
- LiVolsi VA, Abrosimov AA, Bogdanova T, Fadda G, Hunt JL, et al. (2011) The Chernobyl thyroid cancer experience: pathology. Clin Oncol (R Coll Radiol) 23: 261-267. [Crossref]
- Saenko V, Ivanov V, Tsyb A, Bogdanova T, Tronko M, et al. (2011) The Chernobyl accident and its consequences. Clin Oncol (R Coll Radiol) 23: 234-243. [Crossref]
- Schlumberg M, Pacini F (1999) In Thyroid Tumors Schlumberg M, Pacini F (Eds) Nucléon pp.13-30.
- Hsiao SJ, Nikiforov YE (2015) Molecular approaches to thyroid cancer diagnosis. Endrocrine Relat Cancer 21: 1-21.
- Kelly LM, Baril G, Liu P,Evdokimova VN, Trivedi S, et al. (2014) Identification of the transforming STRN-ALK fusion as a potential therapeutic target in the aggressive forms of thyroid cancer. Proc Natl Acad Sci 111: 4233-4238.
- Khan FA, Pandupuspitasari NS, Chun-Jie H, Ao Z, Jamal M, et al. (2016) CRISPR/Cas9 therapeutics: a cure for cancer and other genetic diseases. Oncotarget 7: 52541-52552. [Crossref]
- Dhillon AS, Hagan S, Rath O, Kolch W (2007) MAP kinase signalling pathways in cancer. Oncogene 26: 3279-3290. [Crossref]
- Blais E, Dutertre A (2016) ALK rearrangement in anaplastic thyroid carcinoma: A discovery towards a personalized approach? J Nucl Med Radiat Ther 7: 1-2.
- Mechanick JI, Carpi A (2008) Thyroid cancer: the impact of emerging technologies on clinical practice guidelines. Biomed Pharmacother 62: 554–558.
- Eszlinger M, Paschke R (2010) Molecular fine-needle aspiration biopsy diagnosis of thyroid nodules by tumor specific mutations and gene expression patterns. Mol Cell Endocrinol 322: 29-37.
- Handkiewicz-Junak D, Czarniecka A, Jarzab B (2010) Molecular prognostic markers in papillary and follicular thyroid cancer: Current status and future directions. Mol Cell Endocrinol 322: 8-28.
- Leeman-Neill RJ, Kelly LM, Liu P, Brenner AV, Little MP, et al. (2014) ETV6-NTRK3 is a common chromosomal rearrangement in radiation-associated thyroid cancer. Cancer 120: 799-807.
- Ciampi R, Knauf JA, Kerler R, Gandhi M, Zhu Z, et al. (2005) Oncogenic AKAP9-BRAF fusion is a novel mechanism of MAPK pathway activation in thyroid cancer. J Clin Invest 115: 94-101. [Crossref]
- Pilli T, Prasad KV, Jayarama S, Pacini F, Prabhakar BS (2009) Potential utility and limitations of thyroid cancer cell lines as models for studying thyroid cancer. Thyroid 19: 1333-1342. [Crossref]
- Araldi R, Módolo DG, de Sá Júnior PL, Consonni SR, de Carvalho RF et al. (2016) Genetics and metabolic deregulation following cancer initiation: A world to explore. Biomed Pharmacother 82: 449-458.
- Mittal V (2018) Epithelial Mesenchymal Transition in Tumor Metastasis. Annu Rev Pathol 13: 395-412. [Crossref]
- Santos MJCP, Bastos AU, da Costa VR, Delcelo R, Lindsey SC, et al. (2018) LIMD2 Is Overexpressed in BRAF V600E-Positive Papillary Thyroid Carcinomas and Matched Lymph Node Metastases. Endocr Pathol.
- Cerutti J (2007) Molecular profiling of matched samples identifies biomarkers of papillary thyroid carcinoma lymph node metastasis. Cancer Res 67: 7885-7892.
- Peng H, Farrooji MT, Osborne MJ, Prokop J, McDonald PC, et al. (2014) LIMD2 is a small lim-only protein overexpressed in metastatic lesions that regulates cell motility and tumor progression by directly binding to and activating the integrin-linked kinase. Cancer Res 74: 1390-1403.
- Koch BJ, Ryan JF, Baxevanis AD (2012) The diversification of the lim superclass at the base of the metazoa increased subcellular complexity and promoted multicellular specialization. PLoS One 7: e33261.
- Dawid IB, Breen JJ, Toyama R (1998) LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet 14: 156-162.
- Bach I (2000) The LIM domain: regulation by association. Mech Dev 91: 5-17. [Crossref]
- Feuerstein R, Wang X, Song D, Cooke NE, Liebhaber SA (1994) The LIM/double zinc-finger motif functions as a protein dimerization domain. Proc Natl Acad Sci U S A 91: 10655-10659. [Crossref]
- Kadrmas JL, Beckerle MC (2004) The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol 5: 920-931. [Crossref]
- Pinheiro Dos Santos MJC, Bastos AU, da Costa VR, Delcelo R, Lindsey SC, et al. (2018) LIMD2 Is Overexpressed in BRAF V600E-Positive Papillary Thyroid Carcinomas and Matched Lymph Node Metastases. Endocr Pathol. [Crossref]
- Ferguson J (2017) LIMD2 expression is upregulated in bladder cancer lymph node metastases, correlates with the aggressive basal molecular phenotype, and is implicated in bladder cancer metastasis. J Urol 197: e1314.
- Xing M (2013) Molecular pathogenesis and mechanisms of thyroid cancer. Nat Rev Cancer 13: 184-199. [Crossref]




