Extended-Spectrum β-lactamase-producing Enterobacterales (ESBL-E) constitutes a global burden and is one of the major threats to public health. The production of ESBLs precludes the use of broad-spectrum cephalosporins, making carbapenems the drug of choice for these infections. Thus, the increased prevalence of these organisms has stimulated the empiric use of carbapenems as therapy where ESBL-E are suspected, favoring selection of carbapenem-resistant Enterobacterales. Rapid detection of ESBL-E constitutes a challenge for clinical microbiologists to prevent delaying efficient antibiotic therapy that worsens the survival of the most severely ill patients. Rapid diagnostic tests may identify drug-resistant bacteria, determine antimicrobial susceptibility, and distinguish viral from bacterial infections, therefore guiding effective treatment strategies. Indeed, implementation of strategies for antibiotic stewardship are now required by the Federal government in hospitals across the USA. Rapid diagnostic tests facilitate epidemiological surveillance, by providing the possibility to monitor the transmission of emerging and antibiotic-resistant microorganisms. Traditional techniques for detecting ESBL-PE include phenotypic tests (to detect the presence of the enzyme) or molecular tests (to detect the genes encoding such enzymes). Both current approaches are time-consuming (24-48 h) since they usually require the isolation or growth of the organism before the test is performed. In this review article, we discussed the rapid diagnostic strategies particularly the rapid ESBL test and how such rapid tests can facilitate the surveillance of resistance evolution and guide effective therapeutic strategies.
ESBL, rapid diagnostic, rapid ESBL test, Enterobacterales
In the USA, the number of patients infected with antibiotic-resistant bacteria have reached two-million people each year, and it is estimated that 23,000 of those patients died from bacterial infections not responding to treatment by usual or any antibiotics [1]. Most of the public organizations such as the World Health Organization (2014), the UK Government (2014), the Center for Disease Control and Prevention in the USA (2013) and the Davos Economic Forum in Switzerland (since 2013) have pull the alarm signal to try to control the multidrug resistance. It is estimated that 25,000 patients die each year in Europe due to infections with antibiotic-resistant bacteria [2].
Extended-spectrum β-lactamases (ESBLs) are one of the most prevalent diverse, complex, rapidly emerging resistance mechanisms in Enterobacteriaceae and are considered a serious public health challenge in the US and the rest of the world [3,4]. Those enzymes confer resistance to most β-lactam antibiotics, i.e. penicillin and broad-spectrum cephalosporins (cefotaxime, ceftriaxone, and ceftazidime) with the notable exception of carbapenems. Resistance to β-lactams is a risk factor for therapeutic failure, and, for some special situations, it is a risk factor for death. With regard to empirical therapy, the delayed administration of an effective therapy has a negative effect on the clinical outcome such as the clinical cure, length of hospitalization, or, for the most severe infections or the weakest patients, the survival rate [5]. An estimated 140,000 healthcare associated Enterobacteriaceae infections occur in the United States each year. The Centers for Disease Control and Infection evaluated that ESBL-E are associated with 26,000 drug-resistant infections and 1,700 deaths annually with an excess medical cost for each infection of 40,000 USD in the US alone [6].
Most of the important resistance issues are emerging among Gram negative bacteria which are the main causes of infections for humans and for which very few therapeutic options are left [2]. They are the sources of both community-acquired and hospital-acquired infections (urinary tract infections, septicemia, intra-abdominal infections) [2]. The most clinically significant Gram-negatives in humans belong to the Enterobacterales family, including common nosocomial and community associated pathogens (such as Escherichia coli, Klebsiella spp.) and organisms associated with travel and food-borne diseases (e.g. Salmonella spp.). These bacteria can cause a variety of illnesses ranging from mild urinary tract infections (UTIs) to life-threating conditions including bacteremia and pyelonephritis, leading to septic shock and death. UTIs are among the most prevalent infectious diseases around the world with an estimated overall incidence of 18/1,000 persons per year in the US [7,8]. According to the CDC, UTIs (mostly caused by E. coli) account for more than 8.6 million visits to healthcare professionals each year. Additionally, infections caused by these ESBL-E have been documented in the community, in patients with no important exposure to the healthcare and are of major concern in animal husbandry, suggesting that these organisms can be readily transmitted through the food chain [3].
The biochemical and molecular features of the ESBLs are very distinct since i) they are inhibited by β-lactam inhibitors such as tazobactam and clavulanic acid and the more recent avibactam, ii) ESBL-producers are resistant to most β-lactams including cefotaxime, ceftriaxone, ceftazidime and cefepime with the exception of cephamycins and carbapenems, and ii) ESBL-PE are commonly co-resistant to other classes of antibiotics including aminoglycosides and fluoroquinolones leaving few therapeutic options [3,9]. The most commonly identified ESBLs in Enterobacterales are the CTX-M enzymes, followed by the so- called TEM and SHV derivatives [10].
The prevalence of ESBL producers: adults and children
The prevalence of ESBL producers among clinical isolates of Enterobacterales is highest in Asia (42.0%), followed by the Middle East (37%), Latin America (28%), Europe (18%), Africa (14%), and North America (7.5%) [11]. During 2014, 13–17 out of 22 European countries reported that 85–100% of E. coli and K. pneumoniae isolates were ESBL positive, with highest levels of resistance in eastern countries [12]. The prevalence rate of patients carrying ESBL producers in their gut flora is estimated to be ca. 5% among ambulatory patients attending the cantonal hospital in Geneva [13]. This Swiss rate has been multiplied by 10 in less than ten years but still remains one of the lowest in the world since prevalence rate of ESBL producers may reach from 15% in Paris to 80% in several Asian countries and Latin America [2,3]. In the US, the percentage of healthcare-associated infections caused by ESBL-E has been estimated to be 14% and 23% for E. coli and Klebsiella spp., respectively [6]. Rates of ESBL producers have increased in both E. coli and Klebsiella spp. from ICU patients in the USA, with the most noticeable increase among Klebsiella spp. in Europe [14]. Most importantly, infection by an ESBL-E markedly affect clinical outcomes. Mortality of ESBL-E associated bloodstream infections due to is estimated to be up to 20-25% [15,16]. Studies of risk factors associated with infections by ESBL-E in children focused mainly on hospitalized children [17,18]. Among the American children, the rates of broad-spectrum cephalosporin resistance (G3CR) and ESBL infections in children are increasing in both inpatient and ambulatory settings nationally. During 2014, Out of 368,398 pediatric isolates, 1.97% (7255) were identified as G3CR, and 0.47% (1734) as ESBL producers. The prevalence of both phenotypes increased, respectively, from 1.39% and 0.28% in 1999–2001 to 3% and 0.92% in 2010–2011. The identification of host factors and exposures leading to infection in children such as prior hospitalization, prolonged length of stay, prior antibiotic use, and indwelling devices is essential [19]. In recent years, increasing numbers of children with CA-UTI due to ESBL-E, especially E. coli, have been observed [20]. In the USA, the rates and incidence of ESBL infection are increasing among children younger than 18 years of from 0.53%to 1.4% between January 2003 and December 2007. Isolation of ESBL-producing organisms from young infants (less than 5 months old) presenting to the emergency department or outpatient clinic, likely resulting from contamination from their parents [21]. Additionally, the increased risk of community-acquired ESBL-producing E. coli among children of Middle Eastern ethnic background suggests that a history of international travel or of contact with international travelers is very likely [20]. Topaloglu et al. [22] report that an underlying disease and hospitalization within the last 3 months were risk factors for infection with ESBL-producing E coli and Klebsiella in children [22]. Previous exposure to antibiotics and young age (<1 year) have also been reported to be risk factors [23].
Diagnosing and detecting of ESBL-E
Current techniques for the identification of ESBL producers are based on the phenotypic determination of susceptibility to expanded-spectrum cephalosporins using disk diffusion testing or E-test techniques, followed by the inhibition of the ESBL activity by clavulanic acid or tazobactam. These techniques require the isolation and growth of an organism from the clinical sample resulting in results reported in approximately 48 h [9].
Rapid diagnostic techniques may contribute to the identification of drug-resistant bacteria, determine antimicrobial susceptibility resulting in effective treatment strategies and rapid adaptation of the antibiotic therapy which may save patients’ lives. Indeed, it was demonstrated that the optimization of the antibiotic therapy during the first 6-12 h of infection is crucial for the treatment of life-threatening infections [24].
Molecular methods (including PCR and sequencing) have been developed for the detection of ESBL genes. However, these methods usually require an additional step of bacterial culture (additional 24 h), then 3-8 h to be performed, and require specific and expensive equipment and a significant degree of expertise, apart from the high costs associated [9]. In addition, they detect only known ESBL genes, meaning that any novel emerging resistance gene may be missed until it is formally recognized and identified, and subsequently included in the screening panel.
The biochemical diagnostic for the rapid detection of ESBL activity in Enterobacterales
The identification of broad-spectrum ß-lactamase activity is the cornerstone of the biochemical approach to rapidly detecting broad-spectrum β-lactam resistance. The colorimetric approach for the detection of ESBL activity consists in obtaining a variation in the color of the reagent medium resulting from a hydrolytic activity that modifies the chemical composition of the medium. This variation could be detected by eye.
Advantages of the colorimetric approach
The colorimetric approach is reliable, rapid, cheap, and requires no or very limited additional supplies, fulfilling the requirements of an optimal test for β-lactamase detection. The biochemical detection of ESBL activities allows clinical microbiologists to identify any type of ß-lactamase activity. In addition, one primary advantage of colorimetric approaches is that they can be applied directly to colonies that are grown on selective media for the rapid detection of multidrug-resistant strains.
Disadvantages of the colorimetric approach
The sensitivity of a biochemical test with the aim of detecting an enzymatically mediated mechanism of antibiotic resistance depends on different factors such as (i) the level of expression of the corresponding gene (ii) the ability of the enzyme to hydrolyze the substrate (iii), and the affinity of the enzyme for the substrate
The development of the colorimetric Rapid ESBL NP Test
As discussed before, screening and confirming the presence of an ESBL producer can be technically difficult and is time consuming. This can lead to poor clinical outcome, considering that the time to appropriate antibiotic is crucial in the management of a septic patient [25]. Due to the major diagnostic limitations in detecting ESBL-E, and the major impact in therapy, antibiotic stewardship and patient outcomes, there is an urgent need to develop a reliable, inexpensive and potentially portable approach to rapidly identify ESBL-E. With this rationale, a rapid and cost-effective biochemical test was developed for the detection of ESBL producers within 30 min designated as the Rapid ESBL NP test [10].
This test is based on change in color from red to yellow as a result of hydrolysis of β-lactam ring of broad-spectrum cephalosporin molecule (cefotaxime) generating a carboxyl group into the medium that leads to acidification of the medium and change of color in the sample, which is reversed by addition of tazobactam in positive test (Figure 1). In comparison to a negative control without antibiotic, the reactive tube containing cefotaxime and the pH indicator experiences a color change from red to yellow if ESBL produces some carboxyl-acid groups resulting from cefotaxime hydrolysis. The same reaction is performed in the presence of a penicillinase inhibitor, namely tazobactam, which inhibits the hydrolysis reaction, thereby contributing to identify the ESBL nature of the ß-lactamase. Based on the results of the test, optimal therapeutic and stewardship actions can be potentially implemented. A positive test will indicate the presence of an ESBL-E and might conduct the clinician to use antibiotics that target ESBL. The test may also lead to a rational use of newer β-lactam-β-lactamase combinations (such as ceftolazone-tazobactam or ceftazidime avibactam) according to the local epidemiology of organisms. A negative test will lead for rapid de-escalation and targeted use of therapies which is likely to impact rates of resistance. Of note, results of the Rapid ESBL NP test are obtained within 30 min and are very cheap (our estimates indicate that each test will cost between US$ 3-4). Additionally, the test requires no additional laboratory equipment and can be performed by anyone with minimal training.
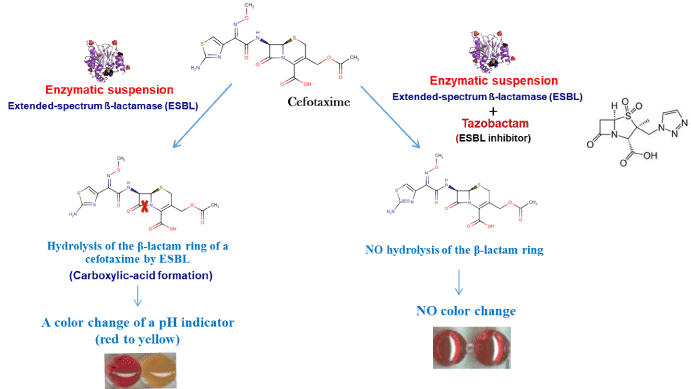
Figure 1. The Rapid ESBL NP test. This test is based on change in colour from red to yellow as a result of hydrolysis of β-lactam ring of cefotaxime generating a carboxyl group that leads to acidification of the medium and change of colour, which is reversed by addition of tazobactam in positive test.
The performance of the Rapid ESBL NP test has been evaluated with either enterobacterial cultured strains (ESBL producers and non-ESBL producers) (sensitivity, 92.6%; specificity, 100%) [10]. Subsequently, 500 urine samples recovered from infected patients (≥104 leukocytes/ml and ≥105 Gram-negative isolates/ml) were investigated for the presence of ESBL producers using the rapid ESBL test. Among the 450 nonduplicate urine samples, 11.3% were positive for ESBL-E. The sensitivity and specificity of the Rapid ESBL NP test were 98% and 99.8%, respectively. A perfect correlation between cefotaxime resistance and positivity of the rapid ESBL NP test was observed [26]. The Rapid ESBL NP test had the ability to detect any type of ESBL especially CTX-M producers such as CTX-M-15 (sensitivity, 100%) which are the most prevalent ESBLs in the US and around the world. ESBL-E were identified prospectively among 245 Gram-negative bacilli-positive cultured blood specimens using the Rapid ESBL NP test. The ESBL test had a sensitivity of 100% (95% CI: 92.4%–100%), a specificity of 100% (95% CI: 97.7%–100%), a positive predictive value of 100% (95% CI: 99.2%–100%) and a negative predictive value of 100% (95% CI: 97.8%–100%) for the detection of ESBL-E [27]. A performance comparison of three tests (the Rapid ESBL NP test (preliminary version termed Rapid ESBL NDP test), the Rapid ESBL Screen kit® which is a copy of the ESBL NP test (Rosco-Diagnostica, Tasstrup, Denmark) and the β-Lacta test®) for detecting the ESBL-producing bacteria was recently performed [28]. The β-Lacta test is based on detection of hydrolysis of a specific extended-spectrum cephalosporin molecule that is chromogenic. The Rapid ESBL NP test reached the best sensitivity and specificity (95% and 100%, respectively). Noteworthy, the β-Lacta test is not specific for detection of ESBL activity, since it also detects production of cephalosporinases (AmpCs) and carbapenemases (such as KPC) that possess an ability to degrade broad-spectrum cephalosporins.
Rapid ESBL NP test using cultured strains
Briefly, strains are isolated from a culture plate before performing the ESBL test. One calibrated inoculated loop of the tested strain is resuspended in lysis buffer. After centrifugation, a mixture of the phenol red solution (with cefotaxime or cefotaxime and tazobactam) and the enzymatic suspension is incubated. The results are read after 30 min. A test result is considered positive when the tube containing cefotaxime alone turns from red to yellow/orange and the well containing cefotaxime supplemented with tazobactam remains red (unchanged color) [10]. This detection of ESBL-E can be performed using cultured bacteria resulting from colonization screening.
Rapid ESBL NP test from positive blood cultures and infected urines
The positive blood culture is centrifuged to pellet the red blood cells, then the supernatant is recovered and centrifuged to pellet the bacteria. For the urine sample, it is centrifuged once to get the bacterial pellet. The bacterial pellet is then resuspended in distilled water with a thorough vortex to lyse the red blood cell remnant and wash the bacterial pellet. The Rapid ESBLNP test is then applied to this pellet as described above (Figure 2). Optical reading of the color change of each tube is used. The ESBL activity is detected through the transformation of cefotaxime into a carboxylic form, leading to a pH decrease revealed by a color change (red to yellow/orange) and inhibition of this reaction by tazobactam leading to no color change [10,27].
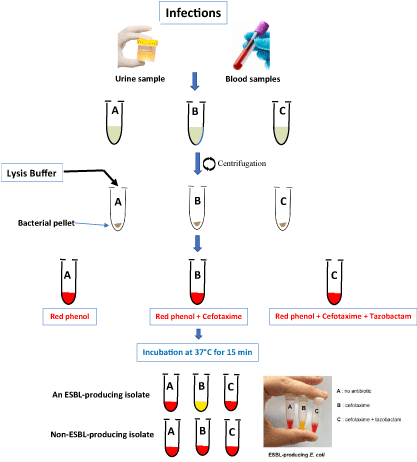
Figure 2. Strategies for applying of the Rapid ESBL NP test to clinical samples (urine and blood samples).
The Rapid ESBL NP test is rapid, sensitive, specific for the early detection of the most prevalent emerging resistance trait, i.e. ESBL-E. It fulfills entirely one of the main identified goals for combating antibiotic resistance that is the development of an innovative diagnostic test for identification and characterization of clinically significant resistant bacteria such as the ESBL producers. Implementation of such test in the strategy of detection of multidrug-resistant bacteria may significantly improve the management and outcome of infected and then colonized patients. In addition, the antibiotic stewardship might be significantly improved leading to the decrease of the selective pressure by over and misuse of carbapenems that plays a crucial role in the emergence and spread of multidrug-resistant bacteria. The Rapid ESBL NP is a biochemical technique that detects any type of ESBL (including the yet unknown) whereas the molecular techniques target only a few of known ESBL genes. As compared to the β-Lacta test that is also a biochemical technique, it selectively detects ESBL activity. Its current industrial development will make it soon a valuable tool for contributing its use worldwide. Finally, the Rapid ESBL NP test is affordable and cost effective compared to molecular techniques. This feature will be of utmost importance particularly for many low resources developing countries where infections with these bacteria constitute a major public health issue.
- Weiner LM, Fridkin SK, Aponte-Torres Z, Avery L, Coffin N, et al. (2016) Vital Signs: Preventing Antibiotic‐Resistant Infections in Hospitals—United States, 2014. Am J Transplant 16: 2224-2230. [Crossref]
- Nordmann P, Poirel L (2014) Emerging and important antibiotic resistance in Gram negative bacteria: epidemiology, theory and practice. Rev Med Suisse 10: 902-907. [Crossref]
- Pitout JDD (2013) Enterobacteriaceae that produce extended-spectrum β-lactamases and AmpC β-lactamases in the community: the tip of the iceberg? Curr Pharm Des 19: 257-263. [Crossref]
- Coque TM, Baquero F, Canton R (2008) Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill 13: 19044. [Crossref]
- Paul M, Shani V, Muchtar E, Kariv G, Robenshtok E, et al. (2010) Systematic review and meta-analysis of the efficacy of appropriate empiric antibiotic therapy for sepsis. Antimicrob Agents Chemother 54: 4851-4863. [Crossref]
- Centers for Disease Control and Prevention (2013) https://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf
- Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, et al. (2001) Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med 29: 1303-1310. [Crossref]
- Hooton TM (2012) Uncomplicated urinary tract infection. N Engl J Med 366: 1028-1037. [Crossref]
- Drieux L, Brossier F, Sougakoff W, Jarlier V (2008) Phenotypic detection of extended‐spectrum β‐lactamase production in Enterobacteriaceae: review and bench guide. Clin Microbiol Infect 14: 90-103. [Crossref]
- Nordmann P, Dortet L, Poirel L (2012) Rapid detection of extended-spectrum-β-lactamase-producing Enterobacteriaceae. J Clin Microbiol 50: 3016-3022. [Crossref]
- Morrissey I, Hackel M, Badal R, Bouchillon S, Hawser S, et al. (2013) A review of ten years of the Study for Monitoring Antimicrobial Resistance Trends (SMART) from 2002 to 2011. Pharmaceuticals 6: 1335-1346. [Crossref]
- European Centre for Disease Prevention and Control (ECDC) (2015) Antimicrobial Resistance Surveillance in Europe 2014. Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net). ECDC, Stockholm.
- Pasricha J, Koessler T, Harbarth S, Schrenzel J, Camus V, et al. (2013) Carriage of extended-spectrum ß-lactamase-producing Enterobacteriacae among internal medicine patients. Antimicrob Resist Infect Control [Crossref]
- Sader HS, Farrell DJ, Flamm RK, Jones RN (2014) Antimicrobial susceptibility of Gram-negative organisms isolated from patients hospitalized in intensive care units in United States and European hospitals (2009–2011). Diagn Microbiol Infect Dis 78: 443-448. [Crossref]
- Schwaber MJ, Carmeli Y (2007) Mortality and delay in effective therapy associated with extended-spectrum β-lactamase production in Enterobacteriaceae bacteraemia: a systematic review and meta-analysis. J Antimicrob Chemother 60: 913-920. [Crossref]
- Tumbarello M, Spanu T, Sanguinetti M, Citton R, Montuori E, et al. (2006) Bloodstream infections caused by extended-spectrum-β-lactamase-producing Klebsiella pneumoniae: risk factors, molecular epidemiology, and clinical outcome. Antimicrob Agents Chemother 50: 498-504. [Crossref]
- Zaoutis TE, Goyal M, Chu JH, Coffin SE, Bell LM, et al. (2005) Risk factors for and outcomes of bloodstream infection caused by extended-spectrum β-lactamase–producing Escherichia coli and Klebsiella species in children. Pediatrics 115: 942-949. [Crossref]
- Kuo K, Shen Y, Hwang K (2007) Clinical implications and risk factors of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae infection in children: a case-control retrospective study in a medical center in southern Taiwan. J Microbiol Immunol Infect 40: 248-254. [Crossref]
- Logan LK, Braykov NP, Weinstein RA, Laxminarayan R (2014) Extended-spectrum β-lactamase–producing and third-generation cephalosporin-resistant Enterobacteriaceae in children: trends in the United States, 1999–2011. J Pediatric Infect Dis Soc 3: 320-328. [Crossref]
- Zhu FH, Rodado MP, Asmar BI, Salimnia H, Thomas R, et al. (2019) Risk factors for community acquired urinary tract infections caused by extended spectrum β-lactamase (ESBL) producing Escherichia coli in children: a case control study. Infect Dis 1-8. [Crossref]
- Blaschke AJ, Korgenski EK, Daly JA, LaFleur B, Pavia AT, et al. (2009) Extended-spectrum β-lactamase-producing pathogens in a children's hospital: A 5-year experience. Am J Infect Control 37: 435-441. [Crossref]
- Topaloglu R, Er I, Dogan BG, Bilginer Y, Ozaltin F, et al. (2010) Risk factors in community-acquired urinary tract infections caused by ESBL-producing bacteria in children. Pediatr Nephrol 25: 919-925. [Crossref]
- Kizilca O, Siraneci R, Yilmaz A, Hatipoglu N, Ozturk E, et al. (2012) Risk factors for community‐acquired urinary tract infection caused by ESBL‐producing bacteria in children. Pediatr Int 54: 858-862. [Crossref]
- Burnham CAD, Leeds J, Nordmann P, O'Grady J, Patel J (2017) Diagnosing antimicrobial resistance. Nat Rev Microbiol 15: 697-700. [Crossref]
- Dhillon RHP, Clark J (2012) ESBLs: a clear and present danger? Crit Care Res Pract (ADD THE CHAPTER AND PAGES) [Crossref]
- Dortet L, Poirel L, Nordmann P (2014) Rapid detection of extended-spectrum-β-lactamase-producing Enterobacteriaceae from urine samples by use of the ESBL NDP test. J Clin Microbiol 52: 3701-3706. [Crossref]
- Dortet L, Poirel L, Nordmann P (2015) Rapid detection of ESBL-producing Enterobacteriaceae in blood cultures. Emerg Infect Dis 21: 504. [Crossref]
- Poirel L, Fernández J, Nordmann P (2016) Comparison of three biochemical tests for rapid detection of extended-spectrum-β-lactamase-producing Enterobacteriaceae. J Clin Microbiol 54: 423-427. [Crossref]


