Protein folding is essential for polypeptides to perform their biomedical functions. Molecular chaperones are known to catalyze protein refolding of tens of thousands of types of polypeptides denatured under stress conditions. However, little is known about the fundamental mechanism of chaperones to recognize and catalyze such an enormous number of species of client polypeptides. Here, I would propose a model for chaperones to associate with their clients by hydrogen bonds between peptide bonds of chaperones and those of client proteins.
There are two types of interactions for protein molecules to associate with other materials, that is, specific and nonspecific interactions. The former is compared to lock and key, while the latter makes it possible for a protein to associate with various partners like a master key. For example, antibodies bind to their antigens in a highly specific manner for protection of our body from invading aliens. On the other hand, major histocompatibility complex binds to any peptide non-specifically for presentation of antigen on the surface of our cells.
Modern medical biology has mainly focused on specific interactions of polypeptides rather than nonspecific ones because specificity of chemical interactions enables living things to develop and maintain their sophisticated organisms by regulating each biochemical reaction individually. Accordingly, biochemists tend to focus on sequence of amino acids of proteins or three-dimension localization of side chains of polypeptides, which give a unique and specific characterization to each polypeptide. In some cases, however, nonspecific binding of polypeptides plays a central role in physiological functions. Antigen-presenting cells such as dendritic cells and macrophages engulf invading microorganisms and degrade alien proteins into short peptides in lysosome. The resultant antigen peptide binds to major histocompatibility complex class II, which subsequently transfers to cell membrane for activation of helper T cells that recognize the antigen peptide specifically. Therefore, major histocompatibility complex should bind to any types of antigen peptide in a sequence-independent manner. Another example of nonspecific binding in our physiological functions is molecular chaperones, which bind to denatured polypeptides to refold them into secondary and tertiary structures properly. Among twenty thousand genes in human genome, molecular chaperones form a family consisting of a limited number of genes including hsp40, hsp70, hsp90, and hsp100. Accordingly, each chaperone should bind to many types of client proteins for maintenance of quality of protein in living cells as a master key.
How is it possible for major histocompatibility complex and molecular chaperones to recognize various peptides in a sequence-independent manner? I would say that they must recognize peptide bonds in primary chain rather than side chain residues of a target peptide. In fact, X-ray crystallography revealed that side chain residues of major histocompatibility complex specifically bind to peptide bonds but not side chain residues of an antigen peptide by hydrogen bonds. This must be the secret of ability for major histocompatibility complex to trap an alien peptide regardless of its amino acid sequence.
How about molecular chaperones? How do they bind to various polypeptides for their proper refolding? Now, we hypothesize that chaperones do not interact with native proteins with proper conformation but interact with polypeptides whose conformation is partially disrupted by heat denaturation. In such a case, it is obvious that chaperones should recognize peptide structures which are exposed outside only after heat denaturation. It might be beta-strands dissociating from beta-barrel or it could be elongating alpha-helices that tend to become random coils or assemble into aggregates of beta-sheets. And as I suggested above, chaperones should associate with denaturing polypeptide via its primary chain rather than side chain residues.
Here, let us simulate refolding process of beta-sheet structure (Figure). Beta-sheet can be stabilized by forming a beta-barrel structure, a cylindrical shape of anti-parallel beta-sheet, as is the case of porins and green fluorescent proteins. Therefore, when anti-parallel strands in a beta-sheet start to separate from each other during heat-induced denaturation, some other beta-sheets could assemble with the beta-sheet to form a barrel-like structure, which stabilize beta-sheet structure of the denaturing beta-sheet. Based on atomic structure, hsp90 dimer appears to be able to stabilize heat-denaturing beta-sheet in client proteins by putting them between two beta-sheets in its client binding domain to form beta-barrel structure as mentioned above. After refolding, the client peptide could be released from hsp90 dimer with ATP hydrolysis, which alters conformation of hsp90 dimer from closed form to open one. This releasing reaction might be similar with that of beta-barrel proteins from SAM50 in mitochondrial outer membrane. It is important to examine the hypothesis that hsp90 protects beta-sheet structure from denaturation, by future experiments.
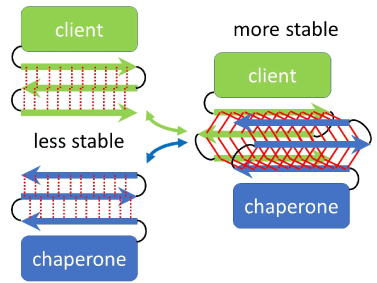
Figure 1. Cartoon of possible molecular mechanism for beta-sheet stabilization of client protein by molecular chaperone; Arrows indicate beta strands; Red lines indicate hydrogen bonds; Dotted and solid lines indicate weak and strong binding, respectively
Taken all together, molecular chaperones may associate with their client by hydrogen bonds between peptide bonds of chaperones and those of client proteins to refold various types of proteins properly in living things.

