Abstract
microRNA are small non coding RNA that target mRNA and regulate various metabolic pathways, cell cycles, etc. Dysregulation of miRNA can lead to serious health issues through translational repression of mRNA and can even cause cancer. In fact, they have a pivotal role in tumorigenesis. Some microRNAs promote tumor formation and thus act as oncogenes while others suppress tumor formation thereby acting as tumor suppressors. microRNAs are involved in many types of cancer, like glioblastoma, leukemia, bladder cancer, pancreatic cancer and gastric cancer. Factors regulating the function of microRNA as oncogenes and tumor suppressors include loss of tumor suppressors and overexpression of oncogenes, the central driving forces for tumorigenesis. The network of oncogene– miRNA–Tumor Suppressor Gene (TSG) affects numerous tumor types and involves several oncogenes, miRNAs, and TSGs; thus contributing in growth and enhancement of a variety of tumors which in turn provides remarkable therapeutic potential for treatment of cancer. Thus understanding the role of microRNA provides opportunities for treatment by providing new drug targets and also acts as a biomarker for diagnostics and therapeutics.
Keywords
microRNA, oncogene, tumor suppressor, cancer, tumorigenesis, therapeutics
Introduction
The microRNAs antagonize target mRNA and are evolutionary conserved, single stranded, "non-coding" RNA molecule. MicroRNAs are transcribed by RNA polymerases II and III, producing precursors that go through a chain of cleavage events to form mature microRNA. These are 19-23 nucleotides long [1]. De-regulation of a miRNA(s) effects the expression pattern of several hundred mRNAs consequently leading to a lot of pathophysiological conditions including cancer [2].
Cancer results due to gene mutations effecting cell proliferation, differentiation and cellular homeostasis. Such genes are grouped as: oncogenes and tumor suppressor genes. The dominant driving forces for tumorigenesis are overexpression of oncogenes and loss of tumor suppressors. Therefore, therapeutic targets of oncogenes and tumor suppressors can give oppurtunities for cancer treatment [3].
Biogenesis of microRNA
The miRNAs transcribed as long primary transcripts, are eventually processed by Drosha and Dicer. Later there is formation of a RNA-inducing silencing complex (RISC) from the mature miRNAs. RISC acts like functional unit which aids in regulation of human gene expression [4]. The miRNAs binds specifically in regions at 3’ UTR of their target mRNA [5] which are involved in a variety of physiological processes.
The primary transcript – primiRNA is transcribed by RNA Polymerase II. The pri-miRNA has single or multiple imperfect hairpin structures with a stem of around 33 base-pairs [6]. The pri-miRNA precursor goes through a two-step processing pathway, mediated by two ribonucleases. The ~ 70 nucleotides long pre-miRNA is cleaved by Drosha, which inturn is sent to the cytoplasm via an exportin-5-dependent mechanism [7].
The pre-miRNA is further processed by Dicer for generating a mature, functional, double-stranded miRNA in the cytoplasm. This is followed by the integration of the guide strand or mature miRNA into a multi-protein complex, RISC, which contains the argonaute (AGO) protein [8]. Along with the guide strand, RISC targets complementary 3′-UTR of mRNA. This is followed by eventual degradation of the other strand also known as miRNA* or passenger strand. The resultant binding of miRNA to the 3′-UTR leads to mRNA degradation or translational repression [9]. However, RISC can also target 5′-UTR of mRNA and activate translation.
Oncomir
An oncomir (oncomiR) is a microRNA that is related with cancer. The oncogene products are grouped as: transcription factors, chromatin remodelers, growth factors, growth factor receptors, signal transducers, and apoptosis regulators [10].
The deregulation of oncomiRs in leukemias and solid cancers and function in cellular gene-reprogramming to establish a phenotypic outcome is not uncommon [11]. The levels of oncomirs vary and is detectable in patient sera upon diagnosis and proposed for diagnostic screening. The miR-372 and miR-373 were found to function as oncogenes in human testicular germ cell tumors. Aberrantly expressed miR 2, an antiapoptotic factor in human glioblastoma cells, is likely to result in malignant human brain cancer [12].
The expression of miR-155 rises up in Burkitt's lymphoma. The oncogenic activity of the BIC gene may be due to miR155 [13], whereas miR-155 over-expression in BIC leads to downregulation of Suppressor of cytokine signaling-1 (SOCS1) as well as activation of the Signal Transducer and Activator of Transcription-3 (STAT3) [14]. The overexpression of miR-155 in BIC downregulates Suppressor of Cytokine Signaling-1 (SOCS1 causing the activation of the Signal Transducer and Activator of Transcription-3 (STAT3) [14].
The emergence of potential new class of targets for therapeutic inhibition has arisen due to the use of miRNAs as biomarkers for targeting and diagnostics [15]. There has been development of several strategies over the recent years for inhibition of oncogenic miRNAs. Of the direct approaches developed to inhibit oncogenic miRNAs, one of them being – targeting mature oncogenic miRNA with an antimiR, either an oligonucleotide or miRNA sponge (direct approach) [16]. On the other hand, genome editing utilizing the CRISPR/Cas9 system or some other small molecule inhibitor is an indirect approach for blocking the biogenesis of miRNA. (
Tumor Suppressor
Tumor suppressor genes cause the inactivation or loss of normal cellular regulatory genes [17]. A multitude of cellular activities are regulated by these genes, like cell cycle checkpoint responses, protein ubiquitylation angiogenesis, cell differentiation and migration, detection and repair of DNA damage, cell cycle checkpoint responses, mitogenic signaling, cell specification, differentiation and migration, protein ubiquitination and degradation, tumour angiogenesis [18].
According to C Stahlhut in 2015,some examples of tumor suppressive miRNA include – let-7, mir-125, miR-339 and miR-766. The miRNA let-7 inhibits lung cancer cell growth and miR-125 has shown its tumor-suppressor functions in several cancers including ovarian cancer, bladder cancer, and breast cancer. The tumor suppressor genes are expressed in colorectal cancer cell lines by miR-339 and 766 via DNA methyltransferase 3B gene inhibition [19].
The tumor-suppressor p53, known for its functions in the DNA damage response and apoptosis [20] and loss or suppression of p53, is a common event in cancers, may act as a significant force in the acquiring the glycolytic phenotype [21,22]. The major regulators of p53 activity as well as its downstream effectors have been studied to be miRNAs. The miR-125b binds conservatively to 3’UTR of p53, whereas miR-504 behaves as a negative regulator of human p53, decreases p53 protein levels, and promotes tumorigenicity. A library screen identified a group of miRNAs regulating p53 activity. Of a miRNA library screen regulating p53 activity, p53 levels are downregulated by miR-30d [23].
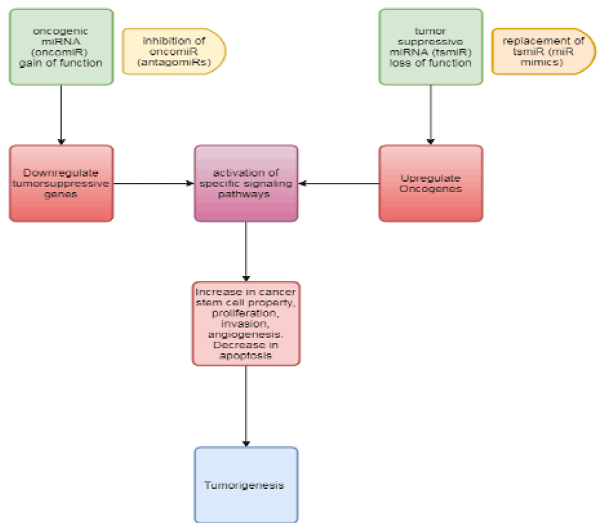
Figure 1. The prevalent, oncogene–miRNA–TSG network in normal cells have important biological functions and leads to various regulatory relationships necessary to keep intermolecular homeostasis for the normal status of cells [32].
Factors regulating microRNA function
RNA binding proteins with double-strand RNA binding domains (dsRBDs), are also known to regulate and control the function of miRNAs along with single nucleotide polymorphism, RNA editing, methylation and circadian clock [24].
A loss-of-function might result in loss of control of miRNA especially those of oncocgenes and drug targets whereas a gain-of-function of an miRNA polymorphism enhance combination of miRNA to the targets, thereby strengthening the regulation effects like tumor suppressor genes [25]. G/C polymorphism within the pre-miR-146a sequence was found to decrease the generation of pre- and mature miR-146a that led to reduction in productive repression of target genes related to the Toll-like receptor and cytokine signaling pathway, which is involved in the genetic susceptibility to papillary thyroid carcinoma [26].
The site-selective alteration of RNA molecules at post-transcriptional level to produce a sequence different from the template DNA is known as RNA editing [27]. It has recently been shown that, the pri-miRNA transcripts of some miRNAs are subject to post-transcriptional modification by A-to-I RNA editing, which is catalyzed by the adenosine deaminases acting on RNA (ADARs) (55–59) (Paul et al., 2017). As in case of miR-142 and miR-151, A-to-I editing at specific positions blocks the Drosha/Dicer cleavage in the maturation of miRNAs [28].
One of the early and frequent events in cancer development has been the aberrant hypermethylation which affects miRNA genes by epigenetic inactivation. Methylation affects the expression of miRNA genes, particularly those located near CpG islands [29]. The over-expression of interleukin-6 leads to downregulation of mir370 while the up-regulation of mir370 is done by the methylation inhibitor 5-aza-2′-deoxycytidine [30].
Network
One of the key players in the complex network of gene regulation are miRNAs and have been involved in various aspects of human disease.
The oncogene– miRNA–TSG network may partake in the development and progression of variety of tumor types, is imbalanced in cancers and can be induced by genetic mutations or epigenetic changes etc., transducer mislead signals to more downstream pathway molecules, and awaken domino effects, therefore, promote the cell malignant transformenmation. The anomaly of this network may be a common event in cancers [31,33].
The oncogene and tumor-suppressor networks influence the metabolic transition in cancer. Furthermore, abnormal cell metabolism and carcinogenesis are largely influenced by the interrelation between deregulated miRNAs and imbalanced signaling pathways. There are many pathways involved in metabolic reprogramming like p53, MYC [33].
Conclusion
The miRNAs can play a dominant role by suppressing oncogenes or tumor suppressors, thereby functioning as tumor-suppressive miRNAs or oncogenic miRNAs during carcinogenesis.
Currently, the change in cell miRNA levels has developed as a potential supportive procedure for an expansive scope of illnesses from hereditary to tumor and viral infection. The overexpression of miRNAs can be decreased utilizing antagomirs, and re-expression of miRNAs that are lost in maladies can be accomplished by the overexpression of miRNA.
Of several miRNAs, miR122 is one miRNA that has successfully reached clinical trials as a targeted therapy. Anti miR-122 could reduce HCV viral load in a chimpanzee model of chronic HCV infection with minimal toxicity [34-42].
References
- Macfarlane LA, Murphy PR (2010) MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics 11: 537-561. [Crossref]
- Sotiropoulou G, Pampalakis G, Lianidou E, Mourelatos Z (2009) Emerging roles of microRNAs as molecular switches in the integrated circuit of the cancer cell. RNA 15: 1443-1461. [Crossref]
- Guo XE, Ngo B, Modrek AS, Lee WH1 (2014) Targeting tumor suppressor networks for cancer therapeutics. Curr Drug Targets 15: 2-16. [Crossref]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, et al. (2004) The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 18: 3016-3027. [Crossref]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science 294: 853-858. [Crossref]
- Quarles KA, Sahu D, Havens MA, Forsyth R, Wostenberg C, et al. (2013) Ensemble Analysis of Primary miRNA Structure Reveals an Extensive Capacity to Deform near the Drosha Cleavage Site. Biochemistry 52: 795-807.
- Zeng Y, Cullen BR (2005) Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. J Biol Chem 28: 27595-27603.
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281-297. [Crossref]
- Walters RW, Bradrick SS, Gromeier M (2010) Poly(A)-binding protein modulates mRNA susceptibility to cap-dependent miRNA-mediated repression. RNA 16: 239-250.
- Pappou, Emmanouil P, Nita A (2010) The Role of Oncogenes in Gastrointestinal Cancer. Gastrointest Cancer Res Suppl 1: S2–S15.
- Giancotti FG (2014) Deregulation of cell signaling in cancer. FEBS Lett 588: 2558-2570. [Crossref]
- Chan JA, Krichevsky AM, Kosik KS (2005) MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 65: 6029-6033. [Crossref]
- Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A (2004) High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer 39: 167-169.
- Mazzarella G, MacDonald TT, Salvati VM, Mulligan P, Pasquale L (2003) Constitutive activation of the signal transducer and activator of transcription pathway in celiac disease lesions. Am J Pathol 162: 845-855.
- Heneghan HM, Miller N, Kerin MJ (2010) MiRNAs as biomarkers and therapeutic targets in cancer. Curr Opin Pharmacol 10: 543-550. [Crossref]
- Nguyen D, Chang S (2018) Development of Novel Therapeutic Agents by Inhibition of Oncogenic MicroRNAs. Int J Mol Sci 19: 65.
- Harris H, Miller OJ, Klein G, Worst P, Tachibana T (1969) Suppression of malignancy by cell fusion. Nature 223: 363-368. [Crossref]
- Sherr CJ (2004) Principles of tumor suppression. Cell 116: 235-246. [Crossref]
- Afgar A, Fard-Esfahani P, Mehrtash A, Azadmanesh K, Khodarahmi F, et al. (2016) MiR-339 and especially miR-766 reactivate the expression of tumor suppressor genes in colorectal cancer cell lines through DNA methyltransferase 3B gene inhibition. Cancer Biol Ther 17: 1126-1138.
- Zilfou JT, Lowe SW (2009) Tumor suppressive functions of p53. Cold Spring Harb Perspect Biol 1: a001883. [Crossref]
- Muller PA, Vousden KH (2014) Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 25: 304-317. [Crossref]
- Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, et al. (2005) Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol 25: 6225-6234.
- Zhang P, Sophie KA, Mohammed S, Elabd S, Heinrich T, et al. (2005) Expression screening using a Medaka cDNA library identifies evolutionarily conserved regulators of the p53/Mdm2 pathway. BMC Biotechnology 15: 92.
- Masliah G, Barraud P, Allain FH (2013) RNA recognition by double-stranded RNA binding domains: a matter of shape and sequence. Cell Mol Life Sci 70: 1875-1895. [Crossref]
- Mishra PJ (2008) MiRSNPs or MiR-polymorphisms, new players in microRNA mediated regulation of the cell: introducing microRNA pharmacogenomics. Cell Cycle 7: 853-858.
- Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, et al. (2008) Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci U S A 105: 7269-7274. [Crossref]
- Barlati S, Barbon A (2005) RNA editing: a molecular mechanism for the fine modulation of neuronal transmission. Acta Neurochir Suppl 93: 53-57. [Crossref]
- Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, et al. (2006) Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol 13: 13-21. [Crossref]
- Lehmann U, Hasemeier B, Christgen M, Müller M, Römermann D, et al. (2008) Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J Pathol 214: 17-24. [Crossref]
- Zhou Y, Hu Z (2015) Genome-wide demethylation by 5-aza-2'-deoxycytidine alters the cell fate of stem/progenitor cells. Stem Cell Rev 11: 87-95. [Crossref]
- Maqbool R, Ismail R, Hussain M (2014) Mutations in MicroRNA Genes and Their Binding Sites are Infrequently Associated with Human Colorectal Cancer in the Kashmiri Population. Microrna 2: 219-224.
- Zhou K, Liu M, Cao Y (2017) New Insight into microRNA Functions in Cancer: Oncogene– microRNA–Tumor Suppressor Gene Network. Front Mol Biosci 4: 46-56.
- Loeb KR, Loeb LA (2000) Significance of multiple mutations in cancer. Carcinogenesis 21: 379-385. [Crossref]
- Jopling CL (2010) Targeting microRNA-122 to Treat Hepatitis C Virus Infection. Viruses 2: 1382-1393. [Crossref]
- Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6: 857-866. [Crossref]
- Cai Y, Yu X, Hu S, Jun Yu (2009) A Brief Review on the Mechanisms of miRNA Regulation. Genomics Proteomics Bioinformatics 7: 147-154.
- Jansson MD, Lund AH (2012) MicroRNA and cancer. Mol Oncol 6: 590-610. [Crossref]
- Kumar M, Lu Z, Takwi AA, Chen W, Callander NS, et al. (2011) Negative regulation of the tumor suppressor p53 gene by microRNAs. Oncogene 30: 843-853. [Crossref]
- Lau NC, Lim LP, Weinstein EG, Bartel DP (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 294: 858-862.
- Paul D, Sinha AN, Ray A, Lal M, Nayak S, et al. (2017) A-to-I editing in human miRNAs is enriched in seed sequence, influenced by sequence contexts and significantly hypoedited in glioblastoma multiforme. Scientific Reports 7: 2466.
- Sengupta S, Harris CC (2005) p53: traffic cop at the crossroads of DNA repair and recombination. Nat Rev Mol Cell Biol 6: 44-55. [Crossref]
- Yimei C, Yu X, Hu S, Yu J (2009) A Brief Review on the Mechanisms of miRNA Regulation. Genomics Proteomics Bioinformatics 7: 147-154.

