Non-small-cell lung cancer (NSCLC) is one of the leading causes of cancer death worldwide. Although recent advances in understanding the pathogenic mechanism of NSCLC has led to the development of targeted treatments, these treatments are only applicable to a limited number of patients. Therefore, the development of a novel therapeutic drug for NSCLC is urgently needed. Here, we focused on miR-301a and miR-301b, belonging to the miR-130 family, because recently, it was reported that miR-130b functions as an oncomiR in NSCLC. The miR-301a and miR-301b were significantly upregulated in NSCLC tissues compared to those in matched-pair adjacent normal lung tissues. Overexpression of miR-301a/b promoted cell proliferation and miR-301b further promoted migration ability in NSCLC cells. Conversely, knockdown of miR-301a and miR-301b significantly suppressed cell proliferation and migration. Moreover, transactivating domain-containing p63 (TAp63), a close relative of the p53 tumor suppressor, was a target gene of both miR-301a and miR-301b in NSCLC cells. These findings showed that miR-301a and miR-301b might function as oncomiRs by targeting TAp63 in NSCLC.
miRNA, non–small-cell lung cancer (NSCLC), oncomiR, miR-130 family
Mortality rate due to lung cancer increases globally every year in Japan and worldwide [1,2]. Lung cancer is histologically classified into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), and the latter accounts for approximately 85% of lung cancer cases. NSCLC is further subdivided into four major histological subtypes: adenocarcinoma, squamous cell carcinoma, large cell carcinoma, and neuroendocrine cancer [3]. Chemotherapy and radiation therapy are generically weakly effective against NSCLC; therefore, surgical removal of cancerous tissue is still the best treatment, despite resulting in low quality of life [4]. In recent years, molecular-targeted drugs, such as gefitinib and erlotinib, have been developed for NSCLC treatment. Although EGFR tyrosine kinase inhibitors have revolutionized the treatment of EGFR-mutated NSCLC, most patients develop acquired resistance via either secondary EGFR mutations or activation of EGFR-independent pathways [5,6]. With five-year survival rate of only 15% for NSCLC, the identification of novel therapeutic targets and development of molecular-targeted drugs is urgently needed.
MicroRNAs (miRNAs) are 20-25 nucleotides long, noncoding RNA molecules. They regulate gene expression through translational repression or mRNA degradation by binding with 3’-untranslated regions (3’-UTR) of a target mRNA [7]. Around 30-60% of human genes are regulated by miRNAs [8,9]; they modulate a variety of cellular processes, such as proliferation, migration, invasion, apoptosis, and angiogenesis [10]. There is no doubt that several miRNAs act as oncogenic miRNAs (oncomiRs) in various cancers [11]. Some miRNAs that share a conserved seed sequence are categorized into a single family and those reported to function in development and progression of cancer are referred to as oncomiRs [12]. Recently, several studies have demonstrated the tumor promoting roles of members of miR-130 family (miR-130b, miR-301a, and miR-301b) [13-15]. For example, miR-130b enhances stem cell-like phenotype in glioblastoma, and miR-301a/b regulates cell proliferation, migration, invasion, and apoptosis in some cancers [16-18]. We previously reported that miR-130 family plays a crucial role in the malignant progression of bladder cancer [19]. Recently, we and others have reported that miR-130b plays an important role in the development and progression of NSCLC [20-22]. However, expression and function of miR-301a and miR-301b in NSCLC remains to be elucidated.
In this study, we elucidated the expression and functional roles of miR-301a and miR-301b in NSCLC specimens and lung cancer cell lines. Our results provided important insights into the molecular pathogenesis of NSCLC and suggested that miR-301a/b may function as oncomiRs in NSCLC.
Clinical specimens
Specimens of NSCLC tissues and adjacent noncancerous tissues were obtained from patients who had undergone primary curative resection of lung tumor at Kagoshima University Hospital (Japan). All enrolled patients were diagnosed by pathological examination and staged by specialized oncologists via the 7th edition of the International Association for the Study of Lung Cancer TNM Classification. A prior written informed consent was obtained from all patients at Kagoshima University Hospital, and the methods were carried out in accordance with approved guidelines. Experiments on clinical specimens were approved by the institutional review boards of Kagoshima University Hospital and the Graduate School of Pharmaceutical Sciences, Osaka University. Clinical and histopathological data related to the specimens are presented in Table 1.
Table 1. Clinical samples used for qRT-PCR analysis
|
Age |
|
Pathological tumor stage |
|
Median |
71 |
T1 |
36 |
Range |
44-88 |
T2 |
11 |
|
|
T3 |
9 |
Gender |
|
Histological subtype |
|
Male |
34 |
Adenocarcinoma |
41 |
Female |
22 |
Squamous carcinoma |
10 |
|
|
Others |
5 |
Quantitative real-time PCR
A miRNeasy Mini kit (Qiagen, MA, USA) was used to isolate miRNA from NSCLC clinical specimens and cancer cell lines. Complementary DNA synthesis was performed using a miR-X miRNA First-Strand Synthesis kit (Takara, Shiga, Japan). Thermal cycler conditions for qRT-PCR included an initial denaturing at 98°C for 30 seconds, followed by 40 cycles of 95°C for 2 seconds and 63°C for 5 seconds, using the following primers: miR-301a, 5'-CAGTGCAATAGTATTGTCAAAGC-3'; miR-301b, 5'-CAGTGCAATGATATTGTCAAAGC-3'; and mRQ 3' primer (Clontech, Freemont, CA, USA). U6 snRNA-specific primers (forward: 5'-TACAAAATACGTGACGTAG-3', reverse: 5'-CTGTTGCTATTATGTCTAC-3') were used as an internal control.
Cell culture
The human lung cancer cell lines A549 and NCI-H520 were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Wako, Osaka, Japan) and RPMI 1640 (Wako), respectively, supplemented with 10% fetal bovine serum (FBS) and 100 mg/mL kanamycin, at 37°C in presence of 5% CO2. A549 and NCI-H520 cells were purchased from Riken cell bank (Tokyo, Japan) and ATCC (Manassas, VA, USA), respectively.
Transfection of miRNA mimics
MiRIDIAN microRNA mimics hsa-miR-301a-3p (C-300657-03-0005), hsa-miR-301b (C-301252-01-0005), and a negative control (CN-001000-01-05) were purchased from GE Healthcare (Lafayette, CO, USA). The mimics were transfected at concentrations of either 10 nmol/L or 50 nmol/L using Lipofectamine 3000 (Life Technologies, Carlsbad, CA, USA), according to the manufacturer’s protocol.
Dual luciferase assay
A pmirGLO dual-luciferase vector was used for the 3'-UTR luciferase reporter assay (Promega, Madison, WI, USA). NCI-H520 cells were co-transfected with a miRNA mimic and a reporter construct containing miR-301a or miR-301b binding site in p63 3'-UTR region. After 36 hours of transfection, dual-luciferase reporter assay was performed according to the manufacturer’s protocol (Promega). Luciferase activity was determined using a luminometer (Turner Biosystems 20/20 luminometer; Promega).
Water-soluble tetrazolium salt-8 (WST-8) cell proliferation assay
After 24 hours of transfection, A549 and NCI-H520 cells were seeded in 96-well plates at 500 and 3000 cells/well, respectively. The cells were incubated at 37°C in presence of 5% CO2, and cell proliferation was examined using Cell Counting kit-8 (Dojin, Kumamoto, Japan) at each time point. The optical density was read at 450/630 nm on a Model 680 Microplate Reader (Bio-Rad, Hercules, CA, USA).
Wound healing assay
After 24 hours of transfection, the cells were seeded at 1×105 cells/well in 24-well plates and cultured to confluence for 24 hours. A wound was created using a 1-mL pipette tip, and images were recorded 0 and 48 hours after wound creation using an Olympus IX71 fluorescence microscope (Olympus, Tokyo, Japan).
Establishment of miR-301a or miR-301b stably knockdown A549 cells
PX330 vectors (Add gene plasmid # 42230) targeting miR-301a and miR-301b were constructed using the following short guide sequences. miR-301a forward: 5'-CCTGCTTTCAGATGCTTTGACAA-3', reverse: 5'-GGACGAAAGTCTACGAAACTGTT-3'; miR-301b (301b-1) forward 1: 5'-AATGATATTGTCAAAGCATCTGG-3', reverse 1: 5'-TTACTATAACAGTTTCGTAGACC-3'; and miR-301b (301b-2) forward 2: 5'-TGGCCGCAGGTGCTCTGACGAGG-3', reverse 2: 5'-ACCGGCGTCCACGAGACTGCTCC-3'. PX330 and a vector containing puromycin resistance gene were co-transfected using Lipofectamine 3000 into A549 cells seeded at 3×104 cells/well in a 12-well plate. After 24 hours of transfection, culture media were exchanged with new media containing 1 mg/mL puromycin. Then the cells were seeded in a 96-well plate (0.7 cells/well) for single clone isolation. The obtained clones were subjected to miRNA purification and subsequent qRT-PCR to confirm downregulated expressions of miR-301a and miR-301b.
Western blot analysis
Whole cell lysates were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to a polyvinylidene difluoride (PVDF: Millipore) membrane by using the semidry transfer system (Bio-Rad, USA). The membranes were probed with specific antibodies, as indicated, and then, incubated with horseradish peroxidase (HRP)-conjugated antibody against mouse or rabbit immunoglobulin (Santa Cruz, USA), followed by detection using enhanced chemiluminescence (ECL) Western blotting detection reagents (GE Healthcare, USA). Image Quant LAS4000 mini system (GE Healthcare) was used as a chemiluminescence detector. The following antibodies were used for immunological analysis in this study: anti-TAp63 antibody (1:2000, BioLegend, 618902) and anti-β-actin polyclonal antibody (1:50000, Sigma-Aldrich, A5316). Densitometric analysis was performed using NIH ImageJ software.
Statistical analysis
Results were expressed as the mean ± S.D. Differences between values were analyzed using a Student’s t-test, Mann-Whitney’s test, or one-way ANOVA with Bonferroni post-hoc tests (GraphPad Prism 5.0, GraphPad Software, San Diego, CA, USA). A p-value < 0.05 was considered statistically significant.
Expressions of miR-301a and miR-301b were significantly upregulated in NSCLC specimens
We first determined the expressions of miR-301a and miR-301b in NSCLC specimens by qRT-PCR. As shown in Figure 1A, the expressions of miR-301a and miR-301b were significantly upregulated in lung cancer tissues compared to adjacent normal lung tissues in 56 matched-pair NSCLC samples. The upregulated expressions of miR-301a and miR-301b were not associated with the stage or the histological type of NSCLC (Figures 1B &1C).
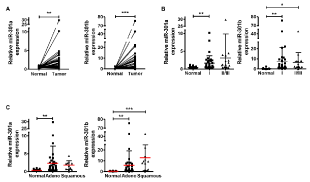
Figure 1. miR-301a/b significantly upregulated in NSCLC specimens
miR-301a/b expression levels were measured by qRT-PCR. Data are represented as relative expression normalized to U6 snRNA. miR-301a/b expressions were compared between normal and tumor tissues (A), among different tumor stages (B), and histological types (C) of NSCLC. Data are presented as mean ± S.D. *p < 0.05, **p < 0.01, ***p < 0.001 for Mann-Whitney’s test (A) and one-way analysis of variance (followed by post-hoc Bonferroni multiple comparison test) (B–C).
miR-301a and miR-301b exhibited tumor-promoting activity in NSCLC cells
To examine the functions of miR-301a and miR-301b, we evaluated their expression levels in HPL1D, a human epithelial cell line derived from normal peripheral lung and NSCLC cell lines. The expressions of miR-301a and miR-301b were found to be downregulated in NCI-H520 cells and upregulated in A549 cells compared to their expressions in HPL1D cells (Supplementary Figure 1). We could confirm the activities of miR-301a/b mimics in NCI-H520 cells using a dual-luciferase reporter assay (Figure 2A & 2B). Then, the effects of the miR-301a and miR-301b mimics on cell proliferation and migration were examined in NCI-H520 cells. Both miR-301a and miR-301b mimics significantly enhanced the proliferation of NCI-H520 cells (Figure 2C). On the other hand, only miR-301b mimic enhanced the migration capability of NCI-H520 cells (Figure 2D).
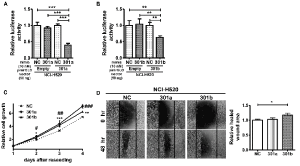
Figure 2. miR-301a/b promoted cell proliferation and migration
miR-301a (A) and miR-301b (B) reporter vectors containing perfectly matched target sites of miR-301a and miR-301b, respectively, were transfected with 10 nM miRNA mimics or a negative control (NC) into NCI-H520 cells. Dual-luciferase reporter assay was performed in the NCI-H520 cells. (C) NCI-H520 cells transfected with miR-301a or miR-301b mimics (50 nM) for 24 h were reseeded in a 96-well plate, incubated for the indicated times and examined by WST-8 assay. (D) NCI-H520 cells transfected with the miR-301a or miR-301b mimics (50 nM) for 24 h were reseeded in a 24-well plate. Relative cell migration was measured 48 h after scratch wound was formed in the cell layer. Data are presented as mean ± S.D. #p < 0.05, ##p < 0.01, ###p < 0.001, *p < 0.05, **p < 0.01, ***p < 0.001 vs. NC for one-way analysis of variance followed by post-hoc Bonferroni multiple comparison test (A-D).
We next carried out loss-of-function experiments using A549 cells, which expressed high levels of miR-301a and miR-301b compared to HPL1D cells. To establish miR-301a- or miR-301b-knockdown A549 cells, we transfected a pX330 vector, targeting miR-301a or miR-301b, into A549 cells. The isolated clones were confirmed to exhibit reduced expression of miR-301a or miR-301b compared to mock vector-transfected A549 cells (Figure 3A). As shown in Figures 3B & 3C, significant suppression of cell proliferation and migration abilities were observed in both miR-301a- and miR-301b-knockdown cells compared to mock vector-transfected A549 cells.
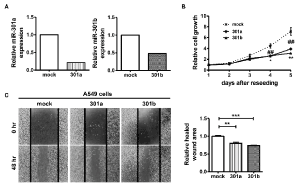
Figure 3.Downregulation of cell proliferation and migration in miR-301a and miR-301b knockdown-A549 cells
(A) miR-301a and miR-301b expressions were quantified by qRT-PCR. (B) Cell proliferation was examined by WST-8 assay. (C) Relative cell migration was measured 48 h after scratch wound was formed in the cell layer. Data are presented as mean ± S.D. of two (A) or three independent (B-C) experiments. ##p < 0.01, *p < 0.05, **p < 0.01, ***p < 0.001 vs. mock cells for one-way analysis of variance followed by post-hoc Bonferroni multiple comparison test (B-C).
miR-301a and miR-301b target TAp63 in NSCLC cells
Finally, to identify a target gene of miR-301a and miR-301b in NSCLC cells, we utilized target prediction programs (miRwalk and TargetScan 7.2) and found that TAp63, a protein isoform of the p63 tumor-suppressor gene, acts as a candidate target gene of miR-301a and miR-301b. To confirm whether TAp63 is a target gene of miR-301a and miR-301b, we transfected a luciferase reporter vector, with a sequence containing the predicted miR-301a/b-binding site (p63 WT) or mutated miR-301a/b-binding site (p63 Mut) within the human p63 3’-UTR, in the NCI-H520 cells (Figure 4A). As shown in Figure 4B, decreased luciferase activity was detected after the transfection of the vector with p63 WT 3'-UTR compared to that of control vector in miR-301a or miR-301b mimic-transfected NCI-H520 cells. On the other hand, miR-301a or miR-301b mimics had no significant effect on luciferase activity of p63 Mut 3'-UTR-transfected NCI-H520 cells (Figure 4B). Moreover, western blot analysis showed decreased TAp63 protein levels in miR-301a/b mimics-transfected NCI-H520 cells (Figure 4C), indicating that TAp63 is a target gene of both miR-301a and miR-301b in NSCLC cells.
To examine the relationship between miR-301a/b and TAp63 protein expression in NSCLC specimens, western blot analysis was performed on NSCLC samples classified into a low or high miR-301a/b expression groups (five samples each group). NSCLC samples with high levels of miR-301a/b expression had significantly lower TAp63 protein expression and vice versa (Figures 4D-4E).
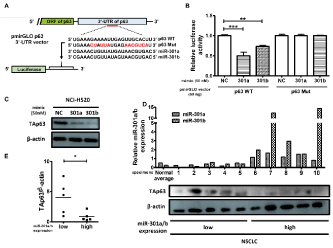
Figure 4. miR-301a and miR-301b target TAp63
(A) The schematic construction of luciferase vector with predicted miR-301a/b-binding sites (p63 WT) and the mutated miR-301a/b-binding sites (p63 Mut) from 3′-UTR of human p63 gene. (B) NCI-H520 cells were co-transfected with a luciferase reporter construct described in (A), along with a miR-301a or miR-301b mimic (50 nM), or the negative control (NC). Values are presented as mean ± S.D. of three independent experiments. **p < 0.01, ***p < 0.001 vs. NC by one-way analysis of variance followed by post-hoc Bonferroni multiple comparison test. (C) miR-301a/b-transfected NCI-H520 cells were subjected to western blot analysis with anti-TAp63 antibody. Representative results of three independent experiments are shown. (D) Taking available amounts of tumor used in Figure 1 into account, five samples of NSCLC tissues with high and five with low miR-301a/b expression were selected. The expression levels of TAp63 and b-actin in the tumor lysates were examined by western blot analysis using their respective antibodies. (E) Densitometric analysis of the western blot was performed using NIH ImageJ and the results were shown as TAp63/b-actin ratio. *p < 0.05, **p < 0.01, ***p < 0.001 for Student’s t-test.
In this study, we showed, for the first time, that miR-301a and miR-301b are overexpressed in NSCLC samples, and function as oncomiRs in NSCLC cells. TAp63, which is a tumor suppressor, was a target gene of miR-301a/b in NSCLC cells. Moreover, expression levels of TAp63 protein were significantly low in NSCLC specimens exhibiting high miR-301a/b expression and vice versa, suggesting that miR-301a/b might function as oncomiRs by targeting TAp63 in NSCLC.
p63 is a close relative of the p53 tumor suppressor protein. p63 gene produces two different protein isoforms: TAp63 that contains a transactivation domain in the N-terminus and functions as tumor suppressor and ∆Np63 that has truncation in the N-terminus and functions as tumor promoter [23,24]. TAp63 activates transcription of several tumor suppressor genes, which, in turn, downregulates cell migration and invasion [25]. Conversely, ∆Np63 acts as an antagonist for TAp63 and p53, which, in turn, inhibits apoptosis [26]. In NSCLC specimens, TAp63 has been previously reported to be downregulated and act as tumor suppressor [27,28]. On the other hand, Lo et al. reported that upregulated ∆Np63:TAp63 expression ratio, rather than single isoform overexpression, showed an association with poor patient outcome in NSCLC [29]. Overexpressed miR-301a and miR-301b might upregulate the ∆Np63:TAp63 expression ratio by downregulating TAp63 expression in NSCLC.
More target gene candidates of miR-301a/b was obtained by miRNA target prediction algorithm databases (miRwalk and TargetScan 7.2), and then, analyzed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (Supplementary Figure 2). Interestingly, Rap1 signaling pathway, Ras signaling pathway, and MAPK signaling pathway were enriched with potential target genes of the miR-301a and miR-301b. It is well known that Rap1 and Ras promote cell proliferation and migration by activating MAPK signaling pathway [30,31]. Our preliminary data also showed that miR-301a and miR-301b mimics increased p-ERK levels but not total-ERK levels in NCI-H520 cells (Supplementary Figure 3). These findings suggested that miR-301a/b might promote cell proliferation by activating MAPK signaling in NSCLC cells.
Rap1 and Ras are “off” in GDP bound state and transduce an “on” signal in GTP bound state. According to miRNA target prediction algorithm databases, Giα, a subunit protein constituting a complex with G-protein-coupled receptor (GPCR), was also a candidate target gene of miR-301a and miR-301b. The Giα subunit activates Rap1 GTPase-activating proteins (Rap1GAPs) and Ras GTPase-activating proteins (RasGAPs), which, in turn, suppress Rap1 and Ras signaling pathway, respectively [32, 33]. Interestingly, candidate target genes of miR-301a and miR-301b were also found among Rap1GAPs (E6TP1) and among RasGAPs (RASA1, RASA2, RASA3, RASA4, RASAL1, RASAL2, and RASAL3). Therefore, miR-301a and miR-301b might constantly activate Rap1 or Ras signaling pathway by downregulating Giα subunit and its downstream targets Rap1GAPs and RasGAPs in NSCLC cells.
There have been several reports showing that miR-130b functions as oncomiR in several cancers [34,35]. Jianwei et al. reported that miR-130b is highly expressed in lung cancer and exhibits an anti-apoptotic effect, suggesting that miR-130b also functions as an oncomiR similar to miR-301a or miR-301b [21]. Taken together, miR-130 family functions as an oncomiR family not only in bladder cancer, but also in NSCLC. We previously reported that miR-130 family-targeted locked-nucleic acid (LNA) significantly inhibits the in vitro and in vivo proliferation of bladder tumor cells, which also overexpress the miR-130 family [36]. Our present study showed that the miR-130 family members function as oncomiRs in NSCLC, suggesting that a nucleic acid drug targeting miR-130 family might act as a promising therapeutic drug against NSCLC.
This study was partially supported by Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED under Grant Number JP18am0101084.
View Supplementary Data
- N Sawabata (2014) Prognosis of lung cancer patients in Japan according to data from the Japanese Joint Committee of Lung Cancer Registry. Respir Investig 52: 317-321.
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, et al. (2015) Global cancer statistics, 2012. CA Cancer J Clin 65: 87-108. [Crossref]
- Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK (2014) Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer 14: 535-546. [Crossref]
- Goldstraw P, Ball D, Jett JR, Le Chevalier T, Lim E, et al. (2011) Non-small-cell lung cancer. Lancet 378: 1727-1740. [Crossref]
- Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, et al. (2005) Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2: e73. [Crossref]
- Yun CH, Mengwasser KE, Toms AV, Woo MS, Greulich H, et al. (2008) The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A 105: 2070-2075. [Crossref]
- Ha M, Kim VN (2014) Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15: 509-524. [Crossref]
- Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215-233. [Crossref]
- Friedman RC, Farh KK, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92-105. [Crossref]
- Esquela-Kerscher A, Slack FJ (2006) Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 6: 259-269. [Crossref]
- Tagawa H, Ikeda S, Sawada K (2013) Role of microRNA in the pathogenesis of malignant lymphoma. Cancer Sci 104: 801-809. [Crossref]
- Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, et al. (2009) miR-19 is a key oncogenic component of mir-17-92. Genes Dev 23: 2839-2849. [Crossref]
- Miao Y, Zheng W, Li N, Su Z, Zhao L, et al. (2017) MicroRNA-130b targets PTEN to mediate drug resistance and proliferation of breast cancer cells via the PI3K/Akt signalling pathway. Sci Rep 7: 41942. [Crossref]
- X Xia, K Zhang, G Cen, T Jiang, J Cao, et al. (2015) MicroRNA-301a-3p promotes pancreatic cancer progression via negative regulation of SMAD4, Oncotarget 6: 21046-21063.
- N Funamizu, CR Lacy, ST Parpart, A Takai, Y Hiyoshi, et al. (2014) MicroRNA-301b promotes cell invasiveness through targeting TP63 in pancreatic carcinoma cells, Int J Oncol 44: 725-734.
- G Zhu, Y Wang, M Mijiti, Z Wang, PF Wu, et al. (2015) Upregulation of miR-130b enhances stem cell-like phenotype in glioblastoma by inactivating the Hippo signalling pathway. Biochem Biophys Res Commun 465: 194-199.
- Cui L, Li Y, Lv X, Li J, Wang X, et al. (2016) Expression of microrna-301a and its functional roles in malignant melanoma. Cell Physiol Biochem 40: 230-244. [Crossref]
- Wu D, Chen B, Cui F, et al. (2016) Hypoxia-induced microRNA-301b regulates apoptosis by targeting Bim in lung cancer. Cell Prolif 49: 476-483. [Crossref]
- Egawa H, Jingushi K, Hirono T, Ueda Y, Kitae K, et al. (2016) The miR-130 family promotes cell migration and invasion in bladder cancer through FAK and Akt phosphorylation by regulating PTEN. Sci Rep 6: 20574. [Crossref]
- Zhang Q, Zhang B, et al. (2018) MicroRNA-130b targets PTEN to induce resistance to cisplatin in lung cancer cells by activating Wnt/β-catenin pathway. Cell Biochem Funct 36: 194-202. [Crossref]
- Tian J, Hu L, Li X, Geng J, et al. (2016) MicroRNA-130b promotes lung cancer progression via PPARγ/VEGF-A/BCL-2-mediated suppression of apoptosis. J Exp Clin Cancer Res 35: 105. [Crossref]
- T Hirono, K Jingushi, T Nagata, M Sato, K Minami, et al., Unpublished data.
- Su X, Chakravarti D, Flores ER (2013) p63 steps into the limelight: crucial roles in the suppression of tumorigenesis and metastasis. Nat Rev Cancer 13: 136-143. [Crossref]
- P Orzol, J Holcakova, M Nekulova, R Nenutil, B Vojtesek, et al. (2015) The diverse oncogenic and tumour suppressor roles of p63 and p73 in cancer: a review by cancer site. Histol Histopathol 30: 503-521. [Crossref]
- Giacobbe A, Compagnone M, Bongiorno-Borbone L, Antonov A, Markert EK, et al. (2016) p63 controls cell migration and invasion by transcriptional regulation of MTSS1. Oncogene 35: 1602-1608. [Crossref]
- Candi E, Dinsdale D, Rufini A, Salomoni P, Knight RA, et al. (2007) TAp63 and DeltaNp63 in cancer and epidermal development. Cell Cycle 6: 274-285. [Crossref]
- HY Zhang, WYang, FS Zheng, Y Wang, JV Lu (2017) Long non-coding RNA SNHG1 regulates zing finger E-box bingidng homeobox 1 expression by interacting with TAp63 and promotes cell metastasis and invasion in Lung squamous cell carcinoma. Biomed Pharmacother 90: 650-658. [Crossref]
- Ko E, Lee BB, Kim Y, Lee EJ, Cho EY, et al. (2013) Association of RASSF1A and p63 with poor recurrence-free survival in node-negative stage I-II non-small cell lung cancer. Clin Cancer Res 19: 1204-1212. [Crossref]
- M Lolacono, V Monica, S Saviozzi, P Ceppi, E Bracco, et al. (2011) p63 and p73 isoform expression in non-small cell lung cancer and corresponding morphological normal lung tissue. J Thorac Oncol 6: 473-481. [Crossref]
- Zhang YL, Wang RC, Cheng K, Ring BZ, Su L, et al. (2017) Roles of Rap1 signalling in tumour cell migration and invasion. Cancer Biol Med 14: 90-99. [Crossref]
- Román M, Baraibar I, López I, Nadal E, Rolfo C, et al. (2018) KRAS oncogene in non-small cell lung cancer: clinical perspectives on the treatment of an old target. Mol Cancer 17: 33. [Crossref]
- DC New, YH Wong (2007) Molecular mechanisms mediating the G protein-coupled receptor regulation of cell cycle progression, J Mol Signal 2: 2.
- P Pamonsinlapatham, R Handj-silmane, Y Lepelletier, B Allain, M Toccafondi, et al. (2009) p120-Ras GTPase activating protein (RasGAP): a multi-interacting protein in downstream signalling. Biochimie 91: 320-328. [Crossref]
- H Chen, Y Yang, J Wang, D Shen, J Zhao, et al. (2018) miR-130b-5p promotes proliferation, migration and invasion of gastric cancer cells via targeting RASAL1. Oncol Left 15: 6361-6367. [Crossref]
- L Satterfield, R Shuck, L Kurenbekova, WA Rhoades, D Edwards, et al. (2017) miR-130b directly targets ARHGAP1 to drive activation of a metastatic CDC42-PAK1-AP1 positive feedback loop in Ewing sarcoma. Int J Caner 141: 2062-2075. [Crossref]
- H Egawa, K Jingushi, T Hirono, R Hirose, Y Nakatsuji, et al. (2016) Pharmacological regulation of bladder cancer by miR-130 family seed-targeting LNA. Integr Mol Med 3: 457-463.




