One of the modern management and organization theory is system approach theory which is consisted of mathematics, biology and genetics. …etc. With the holistic perspective to variety of scientific disciplines have many sustainable relationships between each of them. As regards to numbers and genetic algorithms, system approach is not only seen in golden ratio but also sequenced in nucleotide bases in DNA, too. So at macro level, either in common regulations of many sciences or at micro level in gene expressions, quantum perspective model can be visible. This article is searching for the relation between identical cis regulatory elements and golden mean. Namely, atomic mass structures of TATA Box, CAAT box and GC box relations with golden mean. Researching of TATAA box and CAAT Box gene regulation, the outcome of calculations is approximately very likely to golden mean numbers [1]. Even in one of the step of DNA transcription named as GC Box is also surprisingly unique resemblance to special mathematics number “142”. In sum, pertaining to quantum perspective model at minor level, interrelationship of sciences is ranged from mathematics (Pi and Phi numbers), biochemistry (nucleotide bases and chemical structures), genetics (transcription elements) and also aesthetics in many biological living creatures. At last, the outcome of this article is not only related to golden ratio in mathematics but also nucleotide sequence in cis regulatory elements during transcription in genetics.
golden ratio, system approach theory, GC/AT base pairs, CAAT box, TATAA box
Math. subject classification: 03D45, 97F60, 11R29, 11B75, 92B99, 92E99, QU450, QU34
"General system theory", a synthesis with sciences discipline, mathematics, biology natural and social sciences [2].
According to Giudice, it is estimated that the genomes are closed systems. The role of Phi in the existence of a universal constant, one of the fundamental elements of a complex life code underlying its constitutions, transformations and regulations that mathematical modeling does not have. In principle, science has become broken into interdisciplinary knowledge. Quantum mechanics is the most prominent example. DNA is a closed system. This is related to the fact of the quantitative harmonics that appear to be in the state of DNA. The atomic weights and proteomes of the components is already a result of the open system. All these elements of DNA are actually considered to be the classic particles of codons and multiples in chemical reactions [3].
Gene expression is the process in which genes are copied to different types of RNA (e.g. messenger RNA that leads to protein synthesis). The first step in gene expression activation is defined by the promoter and improves DNA-specific binding of transcription factors [4]. During the gene expression, the effects of cis regulatory elements and brain parts on behavior were investigated [5].
Three stages of transcription: initiation, elongation and termination. While, the first step of this gene regulation is concerned with TATA Box and CAAT Box, the last step of this gene regulation is about GC (G) Guanine and (C) Cytosine / AT (A) Adenine and (T) Thymine base pairs [6].
The region where RNA binds DNA to initiate polymerase transcription is the promoter region. Not all promoters will have the same sequence. Meanwhile powerful promoters will have a similar recurrence sequence as a known consensus sequence. But there will be a different sequence in the set of weak promoters. In the mechanism of gene regulation controlled by cis-regulating elements are listed in the promoter section. Transcription factor is a protein related to regulation of gene expression. It is usually bound to a cis regulatory element. For regulatory proteins to open a gene (activator) or to close (suppressor) to which it is bound to a cis regulatory elements that consist of TATA box and CAAT box [6].
Promoters of human and mouse Claudin-2 genes do not have TATA box during transcription initialization. But the CAAT box indicating that the regulatory elements initiate gene transcription are present (Sakaguchi, 2002). Two hybrid promoters (tacI and tacII) functional in Escherichia coli were produced. The distance between the two domains in tacII is 17 bp (base pairs).In this promoters, it is possible optimal distance to make up for a suboptimal Pribnow box sequence [7]. The root numbers of Phi after comma has an inclusion of about “618” as similar value to cis regulatory elements and base pairs. As seen in calculation of cis regulatory elements (TATA / CAAT Box and GC Box) as a golden ratio (Table 1).From the same quantum perspective, the number of 17 are similiar to tan17.17 (Tan 17, 1717: 0,308) and the value of which is approximately 0,308,too.(Remember,tangent is also ratio,too).As for the same quantum perspective model to double helix of genes,besides previous resemblance, both twin angles of this value “618 “have the same value as after the comma with almost the golden ratio (1, 618034). Approximately, the twin root numbers of Phi is 1,618034 and -0, 618033 [8].
Table 1. Calculation of cis regulatory elements as a golden ratio
Cis regulatory elements and base pairs / Golden Ratio |
Calculation of base pairs (g/mol)
- :70, C:64
- 66, G:78
|
Chemical structures of elements
- :70, C:64
- 66, G:78
|
Phi Value |
GC base pairs (g/mol) |
-
|
-
|
-
|
AT base pairs (g/mol) |
-
|
-
|
-
Lack of One (1) Hydrogen Bond (H:1) |
TATA Box (Average of Nucleotides)
PHOSPHATE GROUPS:
-
SUGAR GROUPS:
- C5H10O5:80
- C5H10O4:72
|
-
Average of sugar phosphate group:136
- 80+50:130
- 72+50:122
- 126
- 482+126:608
|
-
|
-
|
The CAAT Box
|
2*G+3*C+2*A+2*T:620 |
-
|
Extra Two (2) Hydrogen Bond
0.618 |
A rule with numbers is mentioned by pi and phi numbers on the basis of atomic numbers of bases [9]. In sum, after analyzing the results of codon populations, Perez’s genomic numbers are three parameters: 1,2 and Phi number (1,618033…) [10].
There is a significant functional interaction between the CAAT and TATA boxes based on sequence analysis of potential MLPs (Major Late Promoters).These functional transcription elements: The basal TATA box, The distal upstream activating element and The CAAT box [11].
TATA box and GC box
The TATAA box is usually found in nucleotides in the upstream stream from the origin region of the actual transcription. One of the elements associated with eukaryotic RNA support for eukaryotic RNA polymerase II is TATA Box [12].
The eukaryotic polymerase II promoter MLP (Major Late Promoter) contains two basal elements, namely a TATA box and a promoter element [11]. The TATA box is a highly conserved sequence with 24% of human genes in the core promoter. The TATA box has a core DNA sequence 5 '-TATAAA-3'. Generally, 25 base pairs are found in the upstream site during transcription. The TATA binding protein binds to the TATA box. The Pribnow box, like the TATA box, is the first area in which hydrogen bonds between DNA strands are separated. The rich character of the two boxes, Adenine (A) and Thymine (T) allow the separation of Guanine and Cytosine (GC) base pairs because it is easier to separate RNAP II promoter elements of eukaryotes. The initial nucleotide is always Adenine (A), rarely Guanine (G). There is a consensus series called Tata box from the initial site -35 to -25. Next to the Tata box, there are several row boxes positioned at different distances from the TATA box, such as the GC box and CAAT boxes in the upstream zone. These box sequences are transcriptional factors / regulators. Sequences are very crucial for binding of Transcriptional Factors (TF). In the upstream; there are some sequences that can act as binding sites for transcriptional initiation or suppressors that inhibit activator for the activation of transcription [13] (Figure 1).
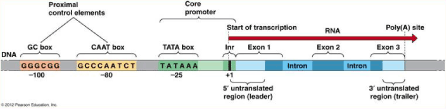
Figure 1. The Common Cis-Regulatory Elements
The CAAT box
The CAAT box is a eukaryotic Basal Transcription Complex (BTC) regulatory element having the 5'-GGCCAATCT-3 'consensus sequence. During the transmission of genetic information, the sequence of CAAT box is GGCCAATCT [14] (Figure 2).
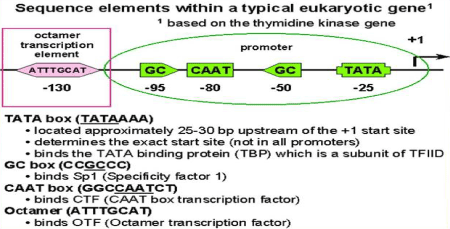
Figure 2. TATA and the CAAT Box, In R (Initiator Region) plus and DPE (Downstream Promoter Element) Containing Promoters
The "main code" is a digital language that combines DNA, RNA and proteins that provides a common alphabet (pi-mass scale) to the three basic languages of genetics, biology and genomics. So, a formula is made with pi-mass projections [9].
From the quantum perspective, not only the expression of Pi number from genetic codes, but also a table which may also apply to the expression of the genetic code of cis regulatory elements as Phi number is shown in Table 1. In addition, the relationship between the two molar masses of AT and GC base pairs and the Golden Ratio value is added to the table below [1,15].
After analyzing of phi numbers versus to cis regulatory elements according to the datum of this research is clarified by both quantum perspective model and other scientific facts for the calculation of the number of phi with genetic codes (Table 2).
Table 2. Representation of DNA nucleotide bases (A, T, C and G) in chemical atoms
ATOMS / NUCLEOTIDE BASES |
C=6 |
H=1 |
O=8 |
N=7 |
SUM |
ADENINE: C5H5N5 |
5 |
5 |
- |
5 |
70 |
THYMINE: C5H6N2O2 |
5 |
6 |
2 |
2 |
66 |
CYTOSINE: C4H5N3O1 |
4 |
5 |
1 |
3 |
64 |
GUANINE: C5H5N5O1 |
5 |
5 |
1 |
5 |
78 |
The atomic numbers of them: Carbon (C):6, Nitrogen (N):7, Oxygen (O):8, Hydrogen (H):1 [16].
The chemical structures of bases are The chemical structure of them [17]:
(A)Adenine: C5H5N5: 70;
(T)Thymine: C5H6N2O2: 66,
(C)Cytosine: C4H5N3O1: 64,
(G)Guanine: C5H5N5O1: 78,
Calculation of cis regulatory elements as a golden ratio
The first one of the cis regulatory elements the TATA box, TATA (T/A) AA [11]. Average value of nucleotides in TATA Box is approximately as similar as value numbers of golden mean after comma (618). The second of the cis regulatory elements is the CAAT box, usually classified as GGCCAAATCT. The relation between golden ratio and the CAAT box is almost similar except for minor difference in calculations [2(two) hydrogen bonds)] (620) (Table 1). The deviation in calculation of the CAAT box can be derived from the Adenine (A) - Thymine (T) hydrogen bonds because Adenine (A) pairs with Thymine (T) by two hydrogen bonds [18]. The number of hydrogen bonds in the bases is two and more than two and this is the common aspect of hydrogen bonds. As a result, the minor difference can be observable in calculations of golden ratio numbers versus to genetic bases. Let alone previous explanations, even the calculation of both AT base pairs and GC base pairs (g/mol) is also surprisingly almost as the same value numbers as golden mean after comma (618). After the comparison of the golden mean values after the comma (618), it can be considered that there is a one lack of hydrogen bond in the AT base pairs (617). The reason of lack of hydrogen bonds: Hydrogen bonding is a very versatile attraction. Hydrogen bonds are relatively weak and easily broken by increasing hardness [12]. It is not only valid in AT pairs but also in GC pairs, too. Finally, the GC box is exactly the same as the third and last 618 (g/mol) value of the GC base pairs of cis regulatory elements and the gold ratio numbers after the exact comma.
The national center for biotechnology information (NCBI) results for cis regulatory elements
The Glycerol-3-phosphate acyltransferase (GPAT) promoters were observed with a plant cis regulatory elements (TATA box, CAAT-box... etc).The GPAT gene promoter is an important regulator under cold stress that improves the resistance of plants for lily. The Lilium pensylvanicum GPAT promoter is a cold source promoter capable of regulating expression at low temperature. As a result, the relationship between promoters and genes has been investigated with GPAT gene promoter [19].
The CAAT box can be the most suitable transcriptional element for special assistance to transcription. According to comparative sequence analysis and the usefulness of a combination of a mutational approach, the conserved CAAT box function is very important for transcription [11]. Surprisingly, after searching triplet times of CAAT Box “GGCCAATCT” in NCBI nucleotide blast, the results are ranged from human beings and mice to others. Likewise, after searching for triple times of TATA Box “TATAAAA” in NCBI nucleotide blast, many types of bony fishes were found, as in CAAT Box. The figures of NCBI nucleotide blast results of TATA and the CAAT Box are specifically focus on variety of bony fishes.
Types of bony fishes are based on CAAT Box: Cyrprinus carpio, Sinocyclocheilus rhhinocerous, Sinocyclocheilus anshuiensis, Atlantic herring, Denticle herring, Yellow catfish, Spiny chromis, Bicolor damselfish and Goldfish.
Types of bony fishes are based on TATA Box: Indian glassy fish, Cottoperca gobio, Blunt-snouted clingfish, Climbing perch, Spotted sea bass, Zig-zag eel, Denticle herring and Takifugu bimaculatus [20].
In sum, for future researchers with a quantum perspective model, more related inferences should be revealed by both genetic sequence or chemical structures and numbers.
At first, the outcomes of this research can be summarized with golden ratio by means of chemical structure of base pairs (AT, CG) and cis regulatory elements. From a narrow perspective, gene sequences can be thought of as phenotypic evolution indicators of beings because they are not only related to genetics and biochemistry at the quantum level, but also aesthetically, they are closely related to other basic sciences such as mathematics, physics and management science. One of the consequences of this evolutionary synthesis is stemmed from varied cis regulatory elements [21]. Changes in the cis regulatory elements contribute to different gene expression, but their contributions are not yet known. Trans-Regulatory differences have influenced more than half of the genes and have always been accompanied by changes in the cis regulatory [22]. Besides, Trans-regulatory regions encoded by transcription factors are very crucial as binding activities of cis regulatory elements. However, functional contributions of these cis elements have not yet been discovered yet. But, even the development and function of neocortex is mainly related to the regulation of gene expression. Therefore, cis regulatory components can be seen in the functioning of many biological organisms [5]. Soon, it may be easy to connect a phenotype change in the gene expression caused by different cis regulatory elements. As a result, these regulatory elements can shed new lights to understand the biological origins of molecular systems [21].
Common feature of NCBI nucleotide blast results of TATA and the CAAT Box is Denticle herring (bony fishes).Interestingly, with a quantum perspective to one of the NCBI result of CAAT Box is goldfish (bony fishes).Even, not only NCBI results for Pi [23] is consisted of types of many bony fishes (Danio Rerio), but NCBI results for Phi also consists of many varied bony fishes, too. The relationships between Pi number’s fourteen groups after comma and genetics codes are explained by [23]. After dividing phi number by fourteen [24]; also it can be possible to reach the cyclic number of “428571” again. This similarity can be regarded as a Quantum Perspective Model [25]. By the same quantum perspective to cis regulatory elements that especially consisting the TATA box, CAAT box and GC/AT base pairs, it is possible to reach the value of golden mean. Finally, at macro level, the result of the evaluation of the aesthetic approach with the ratio of gold attributed to many sciences can be seen not only in social sciences (system approach), biology with NCBI blast results, but also in biochemistry (atomic elements) and genetic sciences (bases, boxes and pairs).At quantum level, Pi numbers are defined as fourteen groups [24] with genetic codes and are similarly defined with an acid base ratio [23]. In addition, the number of "142" can also be associated with golden ratio (Phi), similar to the fact that certain numbers "142857" are related to Pi numbers. Even, the chemical structure of the G-C base pairs is 142 g /mol, too (Table 1). A similar quantum perspective model (such as "142857" number sequence in writing the Pi number as the genetic code) [23] was found in the G-C base pairs with the sequence "142". Both of this value has an inclusion of “14” too. As a result, this aesthetic and dazzling similarity can be the essence of irrational numbers, especially when looking at the cis regulatory elements from a quantum point of view, such as the golden mean values?
- Barbisin M, Sheale JG (2010) Assessment of DNA extracted from forensic samples prior to genotyping. Forensic Science Review 22: 199-214.
- Koçel Tamer (2015) Business Management, İstanbul Kültür University, Beta Yayıncılık pp. 323-324.
- Giudice E Del, Stefani P, Tedeschi A, Vitiello G (2011) The interplay of biomolecules and water at the origin of the active behavior of living organisms, 9th International Froehlich’s Symposium IOP Publishing J Phys Conf Ser 329: 012001.
- Saccani S, Pantano S, Natoli G (2001) Two waves of nuclear factor Kappa B recruitment to target promoters J Exp Med 193: 1351-1359.
- Shibata M, Gulden FO, Sestan N (2015) From trans to cis: Transcriptional regulatory networks in neocortical development. Trends Genet 31: 77-87.
- https://www.ck12.org/book/CK-12-Biology-Advanced-Concepts/section/8.21/
- Herman A (1983) The tac promoter: A Functional Hybrid Derived from the Trp And Lac Promoters” Boer D, Comstock Lisa J and Vasser M. Proceedings of the National Academy of Sciences 80: 21-25.
- Akdeniz Fikri (2007) Golden Mean and Fibonacci Numbers 2007, Nobel Press. Adana pp.9-77
- Perez JC (2015) Deciphering Hidden DNA Meta-Codes -The Great Unification and Master Code of Biology. J Glycomics Lipidomics 5: 131.
- Perez JC (2013) The “3 genomic numbers” discovery: How our genome single-stranded DNA sequence is “Self-Designed” as a numerical whole. Applied Mathematics 2013: 7-53.
- Song B, Young CSH (1998) Functional analysis of the CAAT Box in the Major Late Promoter of the subgroup C Human Adenoviruses, Department of Microbiology, Columbia University, New York, J Virol 72: 3213-3220
- Farrell RE (2010) RNA Methodologies A Laboratory Guide For Isolation and Characterization, 4th Edition, Amsterdam Elsevier Acad Press, The Pennsylvania State University York pp. 704-710.
- http://molbiol4masters.masters.grkraj.org/html/Gene_Structure5BEukaryotic_Promoter_StructureforRNA_Polymerase_II.htm)
- Singh AK, Rana MK, Singh S, Kumar Sundeep, Kumar Rajesh, et al. (2014) CAAT box-derived polymorphism (CBDP): A novel promoter-targeted molecular marker for plants. J Plant Biochem Biotechnol 23: 175–183.
- Nicklas JA, Buel E (2010) Quantification of DNA in forensic samples. Anal Bioanal Chem 376: 1160-1167.
- Wieser EM, Holden N, Coplen BT, Böhlke JK, Berglund M, et al. (2013) Atomic weights of the elements 2011, Pure and Application Chemistry 85: 1047-1078.
- Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, et al. (2018) Molecular Cell Biology, 6th edition, Translation: Geçkil H, Özmen M, Yeşilada Ö, Palme Publishing pp. 294-302.
- http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3353008
- Chen Li-jing, Zhang Li, Qi Wei-kang, Irfan Muhammad, Lin Jing-Wei, et al. (2017) Characterization of the promoter region of the glycerol-3-phosphate-O-acyltransferase gene in Lilium pensylvanicum. Turk J Biol 41: 552-562.
- https://blast.ncbi.nlm.nih.gov/Blast.cgi
- Wittkopp PJ, Kalay G (2012) Cis-regulatory elements: Molecular mechanism and evolutionary process underlying divergence. Nat Rev 13.
- Wittkopp PJ, Haerum BK, Clark AG (2004) Evolutionary changes in cis and trans gene regulation. Nature 430: 85-88.
- Köklü K (2019) A Quantum Perspective Model to Genetic Codes through Various Sciences. Neuroquantology April 17: 3.
- Sakaguchi Takanori, Xiubin Gu, Golden HM, Suh EunRan (2002) Cloning of the human claudin-2 5 flanking region revealed a TATA- less promoter with conserved binding sites in mouse and human for caudal-related homeodomain proteins and hepatocyte nuclear factor-1 alpha. J Biol Chem 277: 24
- Köklü K (2019) Is Relativity Theory Also Valid in Biogenetics and Mathematics? Neuroquantology 17: 3.


