Introduction
Cholesteatoma is a tumorous keratinocytic hyperproliferation with bony destruction and tissue granulation by chronic inflammation. The cholesteatoma that is named as “skin in the wrong place” is the abnormal existence of keratinized squamous epithelium in the middle ear [1]. Cholesteatoma is invasive, hyperproliferative, recidivistic, expansive and destructive [2]. Cholesteatoma is formed by the effect of inflammatory cytokines, growth factors and/or bacterial toxins. The pathogenetic molecular mechanism of the cholesteatoma is still unclear, although it has been studied for a century. Cholesteatoma is thought as a cell growth disorder which involves an altered control of cellular proliferation. However, the reason of this lack of control is still unknown [3]. Cholesteatoma may also be the results of neoplastic transformation event. It is reported that genetic alterations in cholesteatoma are at the gen level not the chromosomal level [2]. A recent study proposed that the gene expression profile of cholesteatoma is similar to a metastatic tumor. In this study, they demonstrated that the cholesteatoma tissue expresses tumor-relevant genes normally expressed in chronically inflamed tissue [4]. In another recent study, several differentially expressed proteins related with cancer were shown in cholesteatoma comparing to corresponding skin lesions. It was reported that these proteins might cause cancer-like alterations and reorganization of the tissue that may explain the characteristic phenotype of the disease [5]. There are not any medical therapies for cholesteatoma and the only applied treatment now is surgical resection. Even after surgery, recurrences are common in cholesteatoma [6]. The aggressive behavior of a cholesteatoma destroying the surrounding bone may cause life threatening complications such as hearing loss, facial palsy, meningitis or intracranial abscesses and this behavior usually is seen in malignant tumor [7]. Thus, there is a need for developing medical treatment alternatives based on the molecular pathology.
The goal of this study was to investigate the genetic instability of the unknown genetic basis of cholesteatoma in human cholesteatoma tissue in comparison to healthy skin using LOH and MSI analysis by capillary electrophoresis via examining some loci in the genome. The studies on microsatellite analysis in most genetic diseases revealed that two major types of alterations could occur: microsatellite instability (MSI) and loss of heterozygosity (LOH). LOH and MSI have been recognized as important events in cancers [8]. Analysis of LOH is an important way to identify informative loci and candidate tumor suppressor genes [9]. Using polymorphic genetic markers, the deletion region can be identified as LOH and also it has shown that LOH is an early event in the progression from normal to neoplastic cells [10]. LOH can be seen in both hereditary and sporadic many genetic diseases, especially tumors, and often it is not known what tumor suppressor genes are, but can be shown as evidence of their existence by LOH [6]. Most studies also show that invasive carcinomas manifest more MSI than precursor lesions, suggesting progressive accumulation of MSI during tumor development [11,12]. With these predictions in mind for cholesteatoma, we have investigated both MSI and LOH.
Materials and methods
Patients and tumor specimens
The study included 14 men and 25 women, with ages ranging from 5 to 77 years (mean; 36.3). 9 of 39 were recurrent; 3 patients had at contralateral ears; 5 of them were primary acquired; 2 of them were congenital and all of the patients had chronic otitis media. The demographic characteristics of the patients are shown in Table 1. Control skin samples were obtained from post auricular surgical incision for each subject with cholesteatoma and also periphery blood samples were obtained during preoperative hematological analysis as a control to determine the status of heterozygosity of specific microsatellite markers. The tissue materials were collected within the framework of scientific and ethical rules from patients in the Department of Otorhinolaryngology between 2010 to 2012. Ethics committee approval required for the study was obtained from the Local Ethics Committee. All samples were stored at −80℃ for DNA extraction.
Table 1. Clinical features of patients in cases studied
| |
Patients |
case number (pediatric/ adult) |
6/29 |
mean of age (pediatric/adult) |
11,2/25,0 |
sex (male/female) |
14/25 |
conjenital/primary acquired/secondary acquired |
3/5/27 |
bilateral/unilateral |
3/35 |
recurrent/nonrecurrent |
9/29 |
DNA extraction
Genomic DNAs were isolated from fresh-frozen tissues and peripheral blood by using kit (Vivantis, Malaysia) according to manufacturer’s orders.
Selection of loci for microsatellite analysis
Three nucleotide repeat markers on four chromosomal arms were used in the study. Primer sequences for the amplification of D6S273, D6S473, D8S261, D9S157, D9S162, D15S126 and D15S153 microsatellite markers are available through the NCBI database (http://www.ncbi.nlm.nih.gov/mapview). All markers of forward primer were fluorescently labeled with 6-FAM (6-carboxyl-fluorescein). The sequences of the primers used are shown in Table 2. Since cholesteatoma is a disease of epithelial origin, markers were selected by reference of a high percentage of LOH in cancers of epithelial origin.
STR |
Primer sequence (5’-3’) |
Localization |
Fragment size (bp) |
Anealing Tm for PCR |
D6S273 |
F: CCCAGTTTAAGGAGTTTGGC
R: TAGGGCAGTCAGCCTTGTG |
6p-6qterm |
120-140 |
56°C for 30 s |
D6S473 |
F: AGCAAGGCAAGCCACAT
R: TGGGGATGCCCAGATAACTATAT |
6p21.3 |
166-196 |
56°C for 30 s |
D8S261 |
F: AGTACCTGAAAGGGTGGG
R: GATCAGTGTAGGCTC |
8p23-p21 |
124-144 |
53°C for 30 s |
D9S157 |
F: AGAACTTGGTATTTCCTGCC
R: GCCACCTTGAGGAGTTTT |
9p23-p11 |
133-149 |
61°C for 30 s |
D9S162 |
F: AGCTGAATAACACGCAGGTG
R: ATTTTTGATGACAATGGAATGC |
9p22 |
172-196 |
63°C for 30 s |
D15S126 |
F: GTAAGCCAAGATGGCACTAC
R: GCCAGCAATAATGGGAAGTT |
15q21 |
188-218 |
61°C for 30 s |
D15S153 |
F: TGCCACTGTCTTGAAAATCC
R: TATGGCCCAGCAATGTGTAT |
15q22.3 |
198-208 |
61°C for 30 s |
Table 2. Sequence, genome localization, size of the product and PCR conditions of STR markers used for LOH analysis are shown
Polymerase chain reactions
PCR reactions were carried out in a 30 µl of reaction mixture containing 0.60 µl of each primer (10 pmol), 4 µl of genomic DNA (up to 50ng), 3 µl of 10×PCR buffer, 0.24 µl of 200 µM of each deoxynucleotide triphosphate, 2 µl of MgCl2 concentrations ranging between 0.75 mM and 2 mM, 0.40 µl of 1 unit of AtMax Taq polymerase and distilled water (dH2O). The reaction mixtures were denatured at 95℃ and incubated 30 cycles, annealing at ranging between 52℃ and 62℃ and an extension step at 72℃ for 1 minute, followed by a final extension step at 72℃ for 7 minutes. The PCR cycles for each marker are given in Table 2. After amplification, 5µl of the reaction mixture was mixed with 2µl of loading dye, and then electrophoresed through 2% agarose gel. The DNA bands were visualized by ethidium bromide staining under ultraviolet light. The concentrations of PCR products were measured by spectroscopy (Thermo Scientific Nanodrop 2000c, USA). These PCR products were diluted with appropriate rates according to their concentrations for LOH and MSI analysis in capillary electrophoresis. All of materials required for gel electrophoresis and PCR were purchased from Vivantis, Malasia.
LOH and MSI analysis
The amplified PCR products were analyzed using the automated ABI Prism sequencer model 310 genetic analyzer (PE Applied Biosystems, USA). 25 μl deionized formamide was combined with 0.5 µl GeneScan-350 (ROX) size standard (PE Applied Biosystems, USA) and 0.5 μl PCR product in a genetic analyzer sample tube. The tubes were closed with genetic analyzer septa and, after short mixing, the samples were denatured in a heat block for 2 minutes at 90℃, chilled on ice, and spin briefly in a microcentrifuge to collect the contents. The samples were loaded on the genetic analyzer.
Results
LOH and MSI analysis
We examined 7 STR markers on chromosomes 6, 8, 9 and 15 in 39 pairs of matched normal and cholesteatoma tissues. Normal and cholesteatoma tissues were evaluated in fragment analysis according to five factors; retension of heterozygosity (ROH), non-informative homozygote, MSI and LOH comparing the peaks in graphics ; 273 pair analysis (run) were performed in total. 60 of them (22%) were not included in the study because of experimental failure of PCR product or LOH analysis. 43 (15.8%) were non-informative. 163 of 273 (59%) were ROH. MSI were present in 7 of 273 (2.6%) and LOH in 1 of 273 (0.4%) (Table 3). LOH was observed in only one patient at D15S153. MSI was observed in one patient at D6S273; two patients at D6S473; one patient at D8S261; two patients at D9S157; one patient at D9S162. There were not any LOH or MSI in any patient at D15S126 (Figure1).
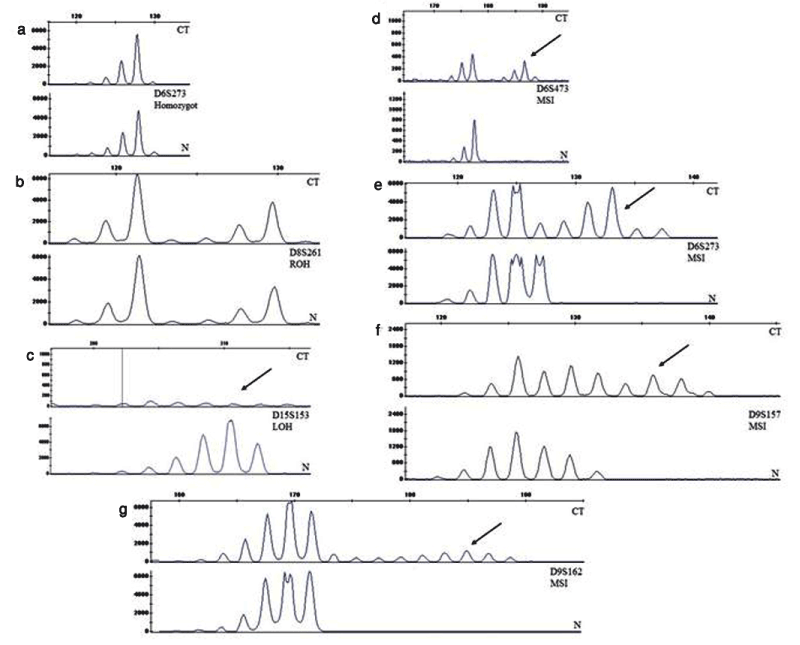
Figure 1. a) Homozygous sample b) Heterozygous sample c) Observed LOH at one mikrosatellite marker D15S153 d-g) Observed MSI at markers D6S273, D6S473, D9S157, D9S162. Electropherograms of examples of selected microsatellite loci. The y-axis represents the peak height in fluorescence units. The arrows indicate an altered allele profile (addition or loss) in the tumor sample compared to the normal DNA. For each marker, the top graph represents cholesteatoma DNA (CT), and the bottom graph represents matching normal DNA (N).
Table 3. Distribution of LOH and MSI for seven microsatellite markers (D6S273, D6S473,D8S261, D9S157, D9S162, D15S126, D15S153) in 39 cases of cholesteatoma
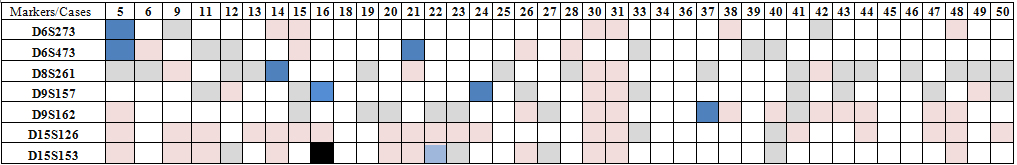
The demographic characteristics of patients showing LOH and MSI
Both LOH and MSI were observed in one patient at age 27 who had secondary acquired cholesteatoma and at contralateral ears. Cholesteatoma in this patient was not recurrent. One patient at age five showed MSI at two STR markers that his cholesteatoma was secondary acquired and bilateral. Only one recurrent cholesteatoma patient had genomic instability. The patients who were at age 50 and 32 presenting MSI had primary acquired cholesteatoma. The other patients presenting MSI had secondary acquired cholesteatoma, and they were 28 and 77 years old. All of these patients had unilateral involvement and no recurrence was observed.
Discussion
Cholesteatoma, described briefly as skin tissue that develops in the wrong place, is a hyperproliferative disorder when epidermal growth control and wound healing process in the middle ear cavity are compromised. Despite numerous studies about molecular biology of cholesteatoma, molecular mechanism of the disease especially the triggering mechanism of hyperproliferation is still not clear. In the present study, we demonstrated the genomes of patients diagnosed with cholesteatoma whether they are genetically stable or not, screening with seven STR markers with the high percentages of LOH in epithelial cancer. In genome-wide screening studies of cholesteatoma, there is controversy about whether it is genetically stable or neoplasm. Bollmann, et al. [13] demonstrated DNA aneuploidy in 9 of 13 patients by interactive cytometry, and they interpreted as a neoplasm. Moreover, other studies reported chromosomal aberrations, such as chromosome 8 aneuploidy [14] and chromosome 7 triploidy that may be associated with hyperproliferation of the cholesteatoma [15]. Also Escence, et al. [16] identified the chromosomal imbalance in the chromosomes of 7, 8 and 17. They reported these aneuploidies to be associated with increased proliferation. In addition to evidence of genetic instability, the MN (micronucleus) frequency, a method for determining chromosome breakage and loss, was also reported to be higher in cholesteatoma tissue than in external ear canal skin [17]. A recent study demonstrated that expression profile of cholesteatoma is similar to a metastatic tumor [4]. Britze, et al. [5] demonstrated that several differentially expressed proteins related with cancer may cause cancer-like alterations and reorganization of the tissue. Taken together, these observations support that cholesteatoma has genetic instability. This is consistent with our results where we demonstrated genomic instability as LOH or MSI in different regions of six patients’ genome.
Frequency of LOH at 8p23-p21 (D8S261) in a larynx squamous cell cancer study was found as 54.5 % and proposed that a tumor suppressor gene is present about at these regions. It was also estimated that this may contribute to transition of preinvasive phase to invasive carcinoma [18]. In our study, MSI was observed in this region in one patient, but LOH was not observed. HLA complex is localized at 6p21.3 and loss of heterozygosity in this region leads to immune escape. For the analysis of this region, D6S273 and D6S473 STR markers were used. Also β2m (beta-2-microglobulin) gene is surrounded by D15S126 and D15S153 STR markers. β2m gene has been associated with HLA class1 expression. Allelic losses of these regions, particularly in many human cancers including head and neck cancers were found in high frequency. It was reported that highest LOH frequency was observed in colorectal and bladder cancers; relatively low LOH frequency was observed in kidney cancer [19]. However, in our study while LOH was not observed in any patients at D6S273 and D6S473 markers, MSI was observed in two patients. LOH of D15S153 was observed in only one patient. In the literature, the loss of 9p21 has been reported to be associated with the risk of head and neck squamous cell cancer relapse [20]. The CDKN2A gene, negative regulator of cell cycle, is localized in this region. Cholesteatoma is also an epithelial origin disease show recurrence. Therefore, D9S157 and D9S162 STR markers covering this region were used in our study. MSI at D9S157 was observed in two patients, and MSI at D9S162 was observed in one patient. In these patients, any recurrence has not been reported. In a study, Hildmann, et al. [21] showed that Ki-67 expression was found higher in pediatric cholesteatoma and reported that cholesteatoma is more aggressive in children. Interestingly, one of our 5 year old patients, who had recurrent cholesteatoma, showed MSI at D6S273 locus.
In conclusion, under the light of the present findings together with other genetic abnormalities reported in the literature, it would be suggested that allelic duplications and losses may be important in cholesteatoma pathogenesis. Therefore, genome-wide screening studies of cholesteatoma will clarify the genetic bases mystery of cholesteatoma.
Acknowledgement
This study was supported by Selcuk University Research Fund (Fund no: 11202029). The authors thank associate professor Ercan KURAR for help and advice with the LOH and MSI analysis.
References
- Gray JD (1964) The Chronic Ear. Proceedings of the Royal Society of Medicine 57: 769.
- Albino AP, Kimmelman CP, Parisier SC (1998) Cholesteatoma: a molecular and cellular puzzle. Am J Otol 19: 7-19. [crossref]
- Derlacki EL, Clemis JD (1965) Congenital cholesteatoma of the middle ear and mastoid. Ann Otol Rhinol Laryngol 74: 706-727. [crossref]
- Klenke C, Janowski S, Borck D, Widera D, Ebmeyer J, et al. (2012) Identification of novel cholesteatoma-related gene expression signatures using full-genome microarrays. PloS one 7: e52718. [crossref]
- Britze A, Birkler RID, Gregersen N, Ovesen T, Palmfeldt J (2014) Large-scale proteomics differentiates cholesteatoma from surrounding tissues and identifies novel proteins related to the pathogenesis. PloS one 9: e104103. [crossref]
- Nussbaum R, McInnes RR, Willard HF (2007) Thompson & Thompson genetics in medicine. Elsevier Health Sciences.
2021 Copyright OAT. All rights reserv
- Huisman MA, de Heer E, Ten Dijke P, Grote JJ (2008) Transforming growth factor beta and wound healing in human cholesteatoma. Laryngoscope 118: 94-98. [crossref]
- Popat S, Hubner R, Houlston R (2005) Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol 23: 609-618. [crossref]
- Peng Z, Zhang F, Zhou C, Ling Y, Bai S, et al. Genome-wide search for loss of heterozygosity in Chinese patients with sporadic colorectal cancer. Int J Gastrointest Cancer 34: 39-47. [crossref]
- Hung J, Kishimoto Y, Sugio K, Virmani A, McIntire DD, et al. (1995) Allele-specific chromosome 3p deletions occur at an early stage in the pathogenesis of lung carcinoma. JAMA 273: 558-563. [crossref]
- El-Naggar AK, Hurr K, Huff V, Clayman GL, Luna MA, et al. (1996) Microsatellite instability in preinvasive and invasive head and neck squamous carcinoma. Am J Pathol 148: 2067-2072. [crossref]
- Ha PK, Pilkington TA, Westra WH, Sciubba J, Sidransky D, et al. (2002) Progression of microsatellite instability from premalignant lesions to tumors of the head and neck. Int J Cancer 102: 615-617. [crossref]
- Bollmann R, Knopp U, Tolsdorff P (1991) DNA cytometric studies of cholesteatoma of the middle ear. HNO 39: 313-314. [crossref]
- Yildirim MS, Ozturk K, Acar H, Arbag H, Ulku CH (2003) Chromosome 8 aneuploidy in acquired cholesteatoma. Acta Otolaryngol 123: 372-376. [crossref]
- Lavezzi A, Mantovani M, Cazzulo A, Turconi P, Matturri L (1998) Significance of trisomy 7 related to PCNA index in cholesteatoma. Am J Otolaryngol 19: 109-112. [crossref]
- Ecsedi S, Rákosy Z, Vízkeleti L, Juhász A, Sziklai I, et al. (2008) Chromosomal imbalances are associated with increased proliferation and might contribute to bone destruction in cholesteatoma. Otolaryngol Head Neck Surg 139: 635-640. [crossref]
- Öztürk K, Yildirim MS, Acar H, ArikoÄźlu H, ArbaÄź H, et al. (2004) Micronucleus frequency in acquired middle ear cholesteatoma. Acta Otolaryngol 124: 924-928. [crossref]
- Yoo WJ, Cho SH, Lee YS, Park GS, Kim MS, et al. (2004) Loss of heterozygosity on chromosomes 3p,8p,9p and 17p in the progression of squamous cell carcinoma of the larynx. J Korean Med Sci 19: 345-351.
- Maleno I, Aptsiauri N, Cabrera T, Gallego A, Paschen A, et al. F(2011) Frequent loss of heterozygosity in the ß2-microglobulin region of chromosome 15 in primary human tumors. Immunogenetics 63: 65-71. [crossref]
- Graveland AP, Golusinski PJ, Buijze M, Douma R, Sons N, et al. (2011) Loss of heterozygosity at 9p and p53 immunopositivity in surgical margins predict local relapse in head and neck squamous cell carcinoma. Int J Cancer 128: 1852-1859. [crossref]
- Hildmann H, Sudhoff H (1999) Cholesteatoma in children. Int J Pediatr Otorhinolaryngol 49 Suppl 1: S81-86. [crossref]


