Phenylenediamine derivatives can function as a hydrogen donor and reportedly exert cytoprotective effects against oxidative stress possibly by acting as an antioxidant. However, our previous report suggested that there is no critical link between the antioxidant ability of these derivatives and prevention of death of neuronal HT22 cells and that some other mechanisms may be involved in protecting neuronal cells against oxidative stress. Here we found that these derivatives might inhibit the cell death pathway by inducing metabotropic glutamate receptor II (mGluRII). By microarray analysis, a phenylenediamine derivative was proved to induce the expression of the mRNA (Grm3) encoding one of the proteins of mGluRII in HT22 cells; and 2S-α-ethylglutamic acid, an antagonist of mGluRII, reduced the protective effects of the derivative. Our results suggest that these compounds protect neuronal cells against oxidative stress, at least in part, by inducing the mGluRII. These compounds may thus be useful tools as mGluRII inducers in neuronal cells.
N, N’-diphenyl-p-phenylenediamine, metabotropic glutamate receptor II, reactive oxygen species, oxidative glutamate toxicity, HT22 cells
DMSO: Dimethyl Sulfoxide; DPPD: N, N’-diphenyl-p-phenylenediamine; ED50: Effective Dose of 50% inhibition; EGLU: (2S)-α-ethylglutamic acid; GSH: Reduced Glutathione; mGluR: Meabotropic Glutamate Receptor; LD50: Lethal Dose for 50% of the cells; MTT: 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide; ROS: Reactive oxygen species; RT-PCR: Reverse Transcription-Polymerase Chain Reaction.
The metabotropic glutamate receptor (mGluR) family greatly expands the potential cellular responses to glutamate within the nervous system [1]. Experiments on the use of selective mGluR agonists and antagonists have shown that these receptors play roles in synaptic plasticity, seizure activity, excitotoxicity, and neuroprotection [2-4]. mGluRII and III are coupled with the Gi and Go families of G-proteins, whereas mGluRI couples to Gq/11 [1]. mGluRI activates phospholipase C (PLC) to generate inositol 1,4,5-triphosphate (IP3) and diacylglycerol, which have multiple “second messenger” roles and finally protect neurons [1]. The activation of mGluRI protects against excitotoxicity and enhances the survival of cultured Purkinje cells, whereas group I antagonists cause retinal degeneration in developing rodents [5].
Phenylenediamine derivatives such as N, N’-diphenyl-p-phenylenediamine (DPPD) exert a cytoprotective effect in vitro against oxidative stress on various types of cells including neuronal cell lines PC12, HT22, and primary cortical neurons [6,7] and inhibit the differentiation of mouse preadipocytes, 3T3-L1 cells, into adipocytes possibly through induction of GDF-15/MIC-1 [8]. DPPD is supposed to protect neuronal cells against oxidative stress at least in part by acting as an antioxidant [7]. However, the neuroprotective effects that we found in a previous study [6] cannot be due to its antioxidant activities in light of the following 3 findings: 1) DPPD does not prevent H2O2-induced cell death; 2) DPPD does not prevent xanthione+xanthine oxidase-induced cell death; and 3) DPPD protects glutamate-treated cells at nM concentrations, which are about 1000 fold lower than those effective to decrease ROS levels [6]. These results suggest that a nM order of DPPD does not function as an antioxidant but that it may inhibit a certain event in the death pathway initiated by oxidative glutamate toxicity [6]. In the present study we addressed straightforwardly this issue; and based on several lines of experimental evidence, we concluded that these compounds at nM levels protected neuronal cells against oxidative stress, at least in part, by inducing the mGluRII.
Compounds
Phenylenediamine derivatives shown in figure 1 were obtained from Ouchi Shinko Chemical Industrial (Tokyo, Japan). D10, D11, D12 and D13 are newly synthesized compounds for the present research. N-Methyl- and N, N-dimethyl-N’-phenyl-p-phenylenediamine (D10 and D11, respectively) were prepared by reductive alkylation of N-phenyl-p-phenylenediamine (PADA) and paraformaldehyde and following chromatographic separation. On the other hand, N-ethyl- and N,N-diethyl-derivative (D12 and D13, respectively) were prepared by direct alkylation of PADA with ethyl bromide. The compounds were dissolved in dimethyl sulfoxide (DMSO). The final concentration of DMSO in the culture medium was 0.1%. Other compounds, including (2S)-α-ethylglutamic acid (EGLU), were purchased from Sigma (St. Louis, MO, USA).
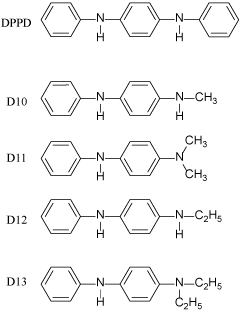
Figure 1: Chemical structures of the phenylenediamine derivatives used in the present study
HT22 culture and MTT assay
HT22 cells were cultured as described previously [9-10]. The cells were maintained in 75-cm2 flasks (Invitrogen, Carlsbad, CA) containing 10 mL of Dulbecco's Modified Eagle medium supplemented with 10% (v/v) heat-inactivated (56°C, 30 min) fetal calf serum (Invitrogen, Carlsbad, CA). For experiments the HT22 cells were seeded into 48-well plates at a density of 4 × 104 cells/cm2. After a 5-h incubation, the desired compounds were added to the cultures. Sixty minutes later, 5 mM glutamate was added; and the cells were then incubated further for 24 h. To evaluate cell survival of HT22 cells, we performed the MTT assay [9-10]. The data were presented as the mean ± SD (for in vitro experiments).
Oligonucleotide microarray analysis
Total RNA was isolated from vehicle (DMSO)- or D11 (10 mM)-treated HT22 cells by using TRIzol Reagent (Invitrogen, Carlsbad, CA) [11-13]. cDNA was synthesized by using the Superscript II system (Invitrogen) with a T7-Oligo(dT) primer. Biotin-labeled cRNA was prepared by in vitro transcription and fragmented by incubation at 94°C for 35 minutes in 40 mmol/L Tris acetate buffer (pH 8.1) containing 100 mM/L potassium acetate and 30 mM/L magnesium acetate. Fragmented cRNA was hybridized at 45°C for 16 hours to a GeneChip® Mouse 430 2.0 Array (Affymetrix, Santa Clara, CA), which contains over 39000 transcripts. Probe arrays were washed and stained using a Fluidics Station 450 and scanned with a GeneChip® Scanner 300. Affymetrix GeneChip Operating Software (GCOS v1.4) was used for analysis.
RT-PCR
Reverse transcription-polymerase chain reaction (RT-PCR) analysis was performed as described previously (9-10, 33-34) using the following primers: for b-actin (287 bp), 5'- ATC CGT AAA GAC CTC TAT GC -3' (forward) and 5'- AAC GCA GCT CAG TAA CAG TC -3' (reverse); and for Grm3 (534bp), 5’-CAAGTGACTACAGAATGCA-3’ (forward) and 5’-CTGTCACCAATGCTCAGCTCT-3’ (reverse).
Protective effects by DPPD derivatives
In HT22 cells and immature cortical neurons, which do not express functional NMDA receptors, high concentrations of glutamate induce a novel type of neuronal death mediated by depletion of reduced glutathione (GSH), termed “oxidative glutamate toxicity” [14,15]. Although these 2 types of neuronal death by glutamate, i.e., excitotoxicity and oxidative glutamate toxicity, are distinct from each other, oxidative stress is involved in both types [14,15], which is why several antioxidant molecules are reported to protect neurons against both types of toxicity [16,17]. Thus, one possible strategy for the development of neuroprotective drugs is to search for low-molecular-weight compounds that can regulate the redox state [18-22]. For the present study, we prepared new lines of DPPD derivatives from D10 to D13 (Figure 1); because DPPD is too hydrophobic and also is reported to be toxic as an allergen and carcinogen possibly through activation of arylhydrocarbon receptors [23]. Thus, we prepared these compounds to break down the structure of one of the benzene rings of DPPD into 1 methyl (D10), 2 methyls (D11), 1 ethyl (D12), and 2 ethyls (D13). By these replacements, at least the hydrophobicity of DPPD must be reduced significantly. At first, we compared the protective effects of these compounds on oxidative glutamate toxicity toward neuronal HT22 cells (Figure 2). In order to assess usefulness of protective compounds against neurodegeneration, we measured the safety zone (=LD50/ED50), as shown in table 1. DPPD was the most potent neuroprotective compound, and it also had the broadest safety zone. D11 is less lipophilic than D1, D13 or DPPD. D11 and D13 had a very low ED50 (70.68 and 17.48 nM, respectively) and had a broad safety zone, and so we selected D11 as a tool of research to investigate the mechanism of the neuroprotective effect of these derivatives.
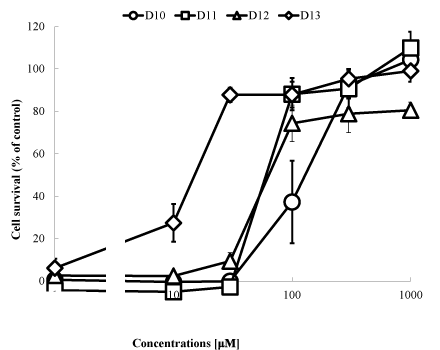
Figure 2: Effects by the compounds on oxidative glutamate toxicity. HT22 cells were seeded into 24-well plates at a density of 4 × 104cells/cm2. After a 5-hr incubation, various concentrations of the compounds were added. One hour later, the cells were treated for 20 h with 5 mM glutamate and then subjected to the MTT assay. The values, which represent the percentage of the control MTT activity, are means + S.D.s (n=4). The experiments presented here were repeated at least 3 times with 4 samples for each determination.
Table 1. Effective dose of 50% inhibition (ED50) of oxidative glutamate toxicity, lethal dose for 50% of the cells (LD50) and safety zone (=LD50/ED50) of each compound.
|
Abbreviation
|
ED50 [nM]
|
LD50[nM]
|
LD50/ED50
|
|
DPPD
|
7.50
|
17544.30
|
2338.67
|
|
D10
|
145.96
|
9813.54
|
67.21
|
|
D11
|
70.68
|
40540.54
|
573.57
|
|
D12
|
73.75
|
7420.60
|
100.61
|
|
D13
|
17.48
|
33713.59
|
1928.49
|
Microarray and RT-PCR analysis
Next, we performed array analysis using D11 in order to identify the gene(s) responsible for the neuroprotective effect by these compounds (Table 2). The cells were incubated with D11 (100 nM) or vehicle, and total RNA was isolated and subjected to reverse-transcription and microarray analysis using the synthesized cDNA. DPPD was earlier reported to activate arylhydrocarbon receptors and to induce cytochrome P450 family genes (Cyp2c55 and Cyp3a41a) significantly [23]. Glutamate receptor metabotropic 3 (grm3) was identified as the gene most induced by this compound, and thus we examined the expression of this gene by RT-PCR (Figure 3A). We observed that Grm3 mRNA was clearly induced by D11 (100 nM), although the levels of b-actin were the same, suggesting that mRNA expression of this gene was really induced by D11 in HT22 cells.
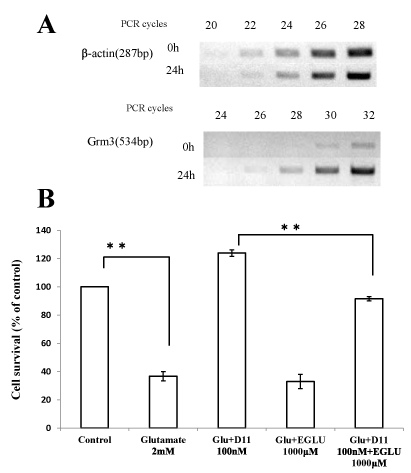
Figure 3: (A) Induction of grm3 gene by D11. Total RNA was extracted from HT22 cells treated with D11 (100 nM) or vehicle (DMSO) for 24 h. RT-PCR was performed for generation and amplification of b-actin and Grm3 cDNAs. Aliquots of the reaction products were subjected to electrophoresis after PCR amplification for the indicated number of cycles. (B) Involvement of mGluR2 in the protective effects afforded by D11. HT22 cells were seeded into 24-well plates at a density of 4 × 104cells/cm2. After a 5-hr incubation, D11 (100 nM) and/or EGLU (1 mM) was added to the cells. One hour later, the cells were incubated for 20 h with/without glutamate 2 mM and then subjected to the MTT assay. The values, which represent the percentage of the control MTT activity, are means + S.D.s (n=4).
Table 2. Top 10 genes induced by D11 in HT22 cells.
|
No.
|
Induction
|
GeneBank
|
Gene description
|
Gene Symbol
|
|
1
|
3.0
|
AK002958
|
Glutamate receptor, metabotropic 3
|
Grm3
|
|
2
|
2.8
|
NM_008331
|
interferon-induced protein with tetratricopeptide repeats 1
|
Ifit1
|
|
3
|
2.6
|
BC008152
|
caspase 1
|
Casp1
|
|
4
|
2.5
|
BC024872
|
receptor transporter protein 4
|
Rtp4
|
|
5
|
2.5
|
NM_021274
|
chemokine (C-X-C motif) ligand 10
|
Cxcl10
|
|
6
|
2.5
|
NM_028089
|
cytochrome P450, family 2, subfamily c, polypeptide 55
|
Cyp2c55
|
|
7
|
2.5
|
AK017218
|
spermatogenesis associated 24
|
Spata24
|
|
8
|
2.1
|
NM_019914
|
myeloid/lymphoid or mixed-lineage leukemia translocated to, 11
|
Mllt11
|
|
9
|
2.1
|
BC012247
|
dicarbonyl L-xylulose reductase
|
Dcxr
|
|
10
|
2.1
|
NM_017396
|
cytochrome P450, family 3, subfamily a, polypeptide 41A
|
Cyp3a41a
|
These genes were selected for their significant up-regulation (p < 0.0003) in response to D11. HT22 cells were incubated in the absence or presence of 100 nM D11 or vehicle for 24 h in normal medium. Total RNA was isolated and subjected to microarray analysis. Fold change (D11/vehicle), GeneBank names, gene descriptions, and gene symbols are indicated.
Pharmacological analysis
Next, we used EGLU, an mGluR2 antagonist, to assess the possible roles of the Grm3 induction in the protective effect of D11. A 24-h incubation of HT22 cells in the presence of 2 mM glutamate resulted in significant cellular degeneration, and 100 nM D11fully protected the cells. Incubation of the cells with both 100 nM D11and 1 mM EGLU decreased the protective effect afforded by D11 alone (Figure 3B). In contrast, EGLU did not affect the viability of the cells (data not shown). These results suggest that the induction of mGluR2 really was involved in the neuroprotective effect mediated by D11.
By the present study, we found that D11, a newly synthesized phenylenediamine derivative, protected HT22 cells against oxidative stress at least possibly through induction of the mGluRII gene. This possibility is supported by the findings that 1) D11 induced expression of the mGluRII gene; and 2) EGLU, a mGluRII antagonist, partially abolished the protective effect afforded by D11. Because HT-22 cells lack ionotropic glutamate receptors and do not constitutively express mGluRII, our results suggest a new type of neuroprotective effect of phenylenediamine derivatives. We propose that diphenylphenylene diamine derivatives can protect neuronal cells possibly through the following 3 mechanisms, 1) antioxidant activities [7,8], 2) activation of arylhydrocarbon receptors [23,24], and 3) induction of mGluRII. Mechanisms 1 and 2 seem to require mM levels of compounds, whereas only mechanism 3 is operative at the nM level. Thus, we can assume that the major pathway contributing to the inhibition of cell death by oxidative glutamate toxicity is the induction of mGluRII. Because these compounds might induce mGluRII in neurons in vivo, they might become effective, novel therapeutics for neuroprotection against neurodegeneration.
The authors thank Dr. Larry D. Frye for editorial help with the manuscript. This work was supported in part by a grant from the JSPS, Joint Project of Japan–U.S. (2008–2010) and by grants to T.S. from the programs Grants-in-aid for Scientific Research (No.19500261, 2007–2009; No. 22500282; 2011-2013 ) and Grants-in-aid for Scientific Research on Innovative Areas (No. 2011701; 2010-2014) of MEXT Japan.
- Nakanishi S (2005) Synaptic mechanisms of the cerebellar cortical network. Trends Neurosci 28: 93-100. [Crossref]
- Flor PJ, Battaglia G, Nicoletti F, Gasparini F, Bruno V (2002) Neuroprotective activity of metabotropic glutamate receptor ligands. Adv Exp Med Biol 513: 197-223. [Crossref]
- Maiese K, Chong ZZ, Shang YC, Hou J (2008) Therapeutic promise and principles: metabotropic glutamate receptors. Oxid Med Cell Longev 1: 1-14. [Crossref]
- Alexander GM, Godwin DW (2006) Metabotropic glutamate receptors as a strategic target for the treatment of epilepsy. Epilepsy Res 71: 1-22. [Crossref]
- Sagara Y, Schubert D (1998) The activation of metabotropic glutamate receptors protects nerve cells from oxidative stress. J Neurosci 18: 6662-6671. [Crossref]
- Satoh T, Izumi M (2007) Neuroprotective effects of phenylenediamine derivatives independent of an antioxidant pathway in neuronal HT22 cells. Neurosci Lett 418: 102-105. [Crossref]
- Satoh T, Sakai N, Enokido Y, Uchiyama Y, Hatanaka H (1996) Survival factor-insensitive generation of reactive oxygen species induced by serum deprivation in neuronal cells. Brain Res 739: 9-14. [Crossref]
- Yanagitai Y, Kitagawa T, Okawa K, Koyama H, Satoh T (2012) Phenylenediamine Derivatives Induce GDF-15/MIC-1 and Inhibit Adipocyte Differentiation of Mouse 3T3-L1 Cells. Biochem Biophys Res Commun 417: 294–298. [Crossref]
- Satoh T, Ishige K, Sagara Y (2004) Protective effects on neuronal cells by ebselen against oxidative stress at multiple steps. Neurosci Lett 16: 1-5. [Crossref]
- Satoh T, Nakatsuka D, Watanababe Y, Nagata N, Kikuchi H, et al. (2000) Neuroprotection by MEK/ERK kinase inhibition against oxidative stress in a mouse neuronal cell line and rat primary cultured neurons. Neurosci Lett 288: 163-166. [Crossref]
- Sasaki S, Tozawa T, Wagoner RM, Ireland CM, Harper MK, et al. (2011) Strongylophorine-8, a pro-electrophilic compound from the marine sponge Petrosia(Strongylophora) corticata, provides neuroprotection through Nrf2/ARE pathway. Biochem Biophys Res Commun 415: 6-10. [Crossref]
- Tamaki Y, Tabuchi T, Takahashi T, Kosaka K, Satoh T (2010) Activated glutathione metabolism participates in protective effects of carnosic acid against oxidative stress in neuronal HT22 cells. Planta Med 76: 683-688. [Crossref]
- Takahashi T, Tabuchi T, Tamaki Y, Kosaka K, Takikawa Y, et al. (2009) Carnosic acid and carnosol inhibit adipocyte differentiation in mouse 3T3-L1 cells through induction of phase 2 enzymes and activation of glutathione metabolism. Biochem Biophys Res Commun 382: 549-554. [Crossref]
- Murphy TH, Schnaar RL, Coyle JT (1990) Immature cortical neurons are uniquely sensitive to glutamate toxicity by inhibition of cystine uptake. FASEB J 4: 1624-1633. [Crossref]
- Dargusch R, Schubert D (2002) Specificity of resistance to oxidative stress. J Neurochem 81: 1394-1400. [Crossref]
- Coyle JT, Puttfarcken P (1993) Oxidative stress, glutamate and neurodegenerative disorders. Science 262: 689–695. [Crossref]
- Satoh T, Lipton SA (2007) Redox regulation of neuronal survival by electrophilic compounds. Trends Neurosci 30: 37-45. [Crossref]
- Satoh T, Okamoto S, Cui J, Watanabe Y, Furuta K, et al. (2006) Activation of the Keap1/Nrf2 pathway for neuroprotection by electrophilic phase II inducers. Proc Nat Acad Sci USA 103: 768-773. [Crossref]
- Satoh T, Raraie T, Seki M, Sunico CR, Tabuchi T, et al. (2011) Dual neuroprotective pathways of a pro-electrophilic compound via HSF-1-activated heat-shock proteins and Nrf2-activated phase 2 antioxidant response enzymes. J Neurochem 119: 569-578. [Crossref]
- Satoh T, Kosaka K, Itoh K, Kobayashi A, Yamamoto M, et al. (2008) Carnosic acid, a catechol-type electrophilic compound, protect neurons both in vitro and in vivo through activation of the Keap1/Nrf2 pathway via S-alkylation of specific cysteines. J Neurochem 104: 1161-1131. [Crossref]
- Davis JB, Maher P (1994) Protein kinase C activation inhibits glutamate-induced cytotoxicity in a neuronal cell line. Brain Res 652: 169–173. [Crossref]
- Sagara Y, Ishige K, Tsai C, Maher P (2002) Tyrphostins protect neuronal cells from oxidative stress. J Biol Chem 277: 36204-36215. [Crossref]
- Sugihara K, Okayama T, Kitamura S, Yamashita K, Yasuda M, et al. (2008) Comparative study of aryl hydrocarbon receptor ligand activities of six chemicals in vitro and vivo. Arch Toxicol 82: 5-11. [Crossref]
- Yeager RL, Reisman SA, Aleksunes LM, Klaassen CD (2009) Introducing the "TCDD-inducible AhR-Nrf2 gene battery." Toxicol Sci 111: 238-246. [Crossref]



