Abstract
Diagnosis of Gleason 6 prostate cancer can leave uncertainty about the presence of undetected aggressive disease. To reduce overdiagnosis and overtreatment of indolent disease, a noninvasive and easy-to-administer test is needed to better stratify risk in patients with clinically insignificant disease. For men on active surveillance, such an assay must also be able to help accurately inform whether a prostate biopsy is warranted, and it must be able to more closely monitor patients with non-aggressive disease.
Objective: To evaluate the performance of a novel, serum-based multiplexed autoantibody assay plus age versus PSA for discriminating high-grade prostate cancer (Gleason Score 7 or greater) from Gleason 6 among patients who have positive biopsies.
Methods: Using retrospective serum samples, the autoantibody assay results were compared with biopsy outcomes in 377 patients (an autoantibody-binding peptide discovery study of 146 samples, a training set of 231 total samples (90 + 141) and a validation cohort comprised of approximately 1/3-2/3 1000X protocol involving a total of 231 samples (90 + 141)) with prostate cancer (Gleason 6 or greater). Samples were sourced from two academic sites and one community clinical facility in the United States. Eligible participants included men aged 40 years or older undergoing a prostate biopsy due to suspicious digital rectal examination finding and/or elevated PSA level.
Results: Among the 377 men (median age 63.2 years; median PSA 7.4 ng/mL), the autoantibody assay plus Age showed better discrimination (Area Under the Curve (AUC) 0.83, 95% CI 0.76-0.90) than PSA alone (AUC 0.60, 95% CI 0.52-0.68) (P =0.001) for separating Gleason Score 7 or greater from Gleason Score 6 in prostate cancer positive patients. For detected Gleason Score 7 or higher test with 95% sensitivity, 24.4% of total biopsies could have been avoided, missing only 3.9% or six (6) patients with Gleason Score 7 disease. Of these six (6) patients whose cancers were missed, a sub-analysis revealed five were Gleason 7 (3+4) and only one was Gleason 7 (4+3).
Conclusion: This study evaluated a novel, cancer-specific biomarker assay based on autoantibody signatures that could be used as a noninvasive risk assessment aid for men with non-aggressive disease who are not receiving treatment. When results of this assay are combined with traditional clinical risk factors, risk stratification for high-grade prostate cancer and biopsy decision making are improved compared to current methods in clinical practice. We hypothesize patients with Gleason Score 6 will exhibit the greatest benefits from employing the assay as they may likely avoid both unnecessary prostate biopsy and subsequent overtreatment. We further hypothesize the assay will thus significantly reduce costs to the healthcare system while further improving patient’s quality of care. Providers and their patients diagnosed with indolent prostate cancer may consider using this novel assay prior to proceeding with prostate biopsy or other treatments.
Introduction
Low risk and many cases of low-intermediate risk prostate cancer are indolent, have little or no metastatic potential, and do not pose a threat to the patient in his lifetime [1]. Active Surveillance is increasingly accepted as a treatment option for favorable-risk prostate cancer as it may reduce the risk of overtreatment of clinically insignificant prostate cancer while retaining the option for definitive therapy for patients who are reclassified over time as higher risk [2].
Active surveillance protocols have evolved in recent years. There are variations between the protocols of different institutions, and a standardized protocol does not yet exist. The principle modality of active surveillance follow-up is a combination of serial prostate-specific antigen (PSA) measurements and digital rectal exams (DRE). An unmet need among physicians and patients using the active surveillance approach has been a means of avoiding the 25% to 30% risk of misclassification inherent in a systematic biopsy-based diagnostic strategy [1].
Given variation among the recommendations and associated controversy with PSA-based prostate cancer screening, clinicians are encouraged to balance harms and benefits from screening to prevent negative physical and psychological side effects from diagnosis and therapy [3]. Whilst PSA remains the first-line biomarker option for the detection of prostate cancer, significant research efforts on novel serum, urine and tissue biomarkers to identify patients at risk for intermediate- or high-risk prostate cancer are underway. However, unlike the serum-based novel multiplexed autoantibody assay described herein, many of these other biomarkers are based in some way on measurement of PSA [4].
Following a prostate cancer diagnosis, patients are faced with a multitude of care options, the advisability of which is influenced by patient factors and by cancer severity or aggressiveness. The ability to categorize patients based on cancer aggressiveness is invaluable for facilitating care decisions. Recently, the American Urological Association (AUA) in conjunction with the American Society for Radiation Oncology (ASTRO) and the Society of Urologic Oncology (SUO) issued guidelines recommending active surveillance for patients for very low-risk and most low-risk localized prostate cancer patients [5]. That said, a novel autoantibody assay that can assist in the assessment of cancer severity and subsequent risk group would likely be clinically useful.
Building on the advances in genomics and proteomics, and specifically the discovery that patients with cancer produce detectable autoantibodies against antigens in their tumors, suggests that these biomarkers could have diagnostic and prognostic value [6]. Researchers have identified a panel of eight peptides that are recognized by autoantibodies generated by the immune system in response to the presence of prostate cancer, and developed an algorithm that can be used to indicate a relatively high or low risk of prostate cancer, particularly for patients with intermediate (4.0 to 10 ng/mL) PSA levels [4].
These cancer-specific, non-PSA blood tests - known as Apifiny® and Apifiny® PRO - in a variety of clinical studies [4,7] have provided insights into the presence and aggressive prostate cancer risk in patients being considered for initial or repeat biopsy.
The aim of this study was to validate the performance of this expanded serum-based multiplexed autoantibody assay in the risk stratification of men with non-aggressive disease who are not receiving treatment. Thus, researchers examined the performance of the 8 Apifiny® peptides when used in combination with an extended set of up to 13 new autoantibody-binding peptides plus age compared to PSA in discriminating high-grade prostate cancer (Gleason Score 7 or greater) from Gleason 6 among patients who have positive biopsies [8].
Materials and methods
Study population
Retrospective serum samples and annotated clinical information for 377 biopsy positive patients were sourced from two academic sites and one community facility in the United States. Eligible participants included men aged 40 years or older, undergoing a prostate biopsy due to suspicious digital rectal examination finding and/or elevated PSA level. Only men with previously diagnosed prostate cancer were excluded from the study.
The men had an average age of 63.2 and an average PSA of 7.4ng/mL. Seventeen percent of men were aged 40-55, 49% were between the ages of 55 to 65, 24% were aged 65-75 and 10% of men were older than 75 years. Twenty-two percent of men had a PSA <4.0 ng/mL, 64% had a PSA between 4 to 10 ng/mL, and only 14% of men had a PSA greater than 10 ng/mL. The majority (83%) of the men were Caucasian, 10% were African American and 7% were classified as ‘other’, likely due to the fact that their race was not documented. Twenty-seven percent of men had Gleason 6 cancer, 30% had Gleason 7 (3+4) cancer, 7% had Gleason 7 (4+3) cancer, and 19% of men had Gleason 7 cancer with no breakout. Finally, 17% of men had Gleason 8 or greater. Because all samples were from men with biopsy- confirmed prostate cancer, almost all subjects (98%) had only one biopsy. Put another way, only 2% or three (3) men in the validation cohort underwent a repeat biopsy (Tables 1-5).
Table 1: Age Ranges from Samples in the Training and Validation Studies
AGE Ranges |
Training Samples |
Validation Samples |
TOTAL |
40-55 |
17 |
19% |
40-55 |
22 |
16% |
39 |
17% |
> 55 to 65 |
43 |
48% |
> 55 to 65 |
70 |
50% |
113 |
49% |
> 65 to 75 |
20 |
22% |
> 65 to 75 |
35 |
25% |
55 |
24% |
> 75 |
10 |
11% |
> 75 |
14 |
10% |
24 |
10% |
TOTAL |
90 |
|
TOTAL |
141 |
|
231 |
|
AVG Age |
63.2 |
|
AVG Age |
63.1 |
|
|
|
Table 2: PSA Levels from Samples in the Training and Validation Studies
PSA Levels |
Training Samples |
Validation Samples |
TOTAL |
less than 4 |
18 |
20% |
less than 4 |
33 |
23% |
51 |
22% |
4 to 10 |
57 |
63% |
4 to 10 |
90 |
64% |
147 |
64% |
> 10 |
15 |
17% |
> 10 |
18 |
13% |
33 |
14% |
TOTAL |
90 |
|
TOTAL |
141 |
|
231 |
|
AVG PSA |
8.15 |
|
AVG PSA |
6.7 |
|
|
|
Table 3: Race Breakout from Samples in the Training and Validation Studies
Race Breakout |
Training Samples |
Validation Samples |
Total |
African American |
8 |
9% |
African American |
15 |
11% |
23 |
10% |
Caucasian |
76 |
84% |
Caucasian |
116 |
82% |
192 |
83% |
Other |
6 |
7% |
Other |
10 |
7% |
16 |
7% |
Total |
90 |
|
Total |
141 |
|
231 |
|
Table 4: Gleason Scores from Samples in the Training and Validation Studies
Gleason Score Breakout |
Training Samples |
Validation Samples |
Six |
26 |
29% |
Six |
38 |
27% |
Seven (3+4) |
27 |
30% |
Seven (3+4) |
42 |
30% |
Seven (4+3) |
6 |
7% |
Seven (4+3) |
10 |
7% |
Seven (w/out breakdown) |
13 |
14% |
Seven (w/out breakdown) |
27 |
19% |
Eight |
7 |
8% |
Eight |
7 |
5% |
Nine + |
11 |
12% |
Nine + |
17 |
12% |
TOTAL |
90 |
100% |
TOTAL |
141 |
100% |
Table 5: Number of Biopsies from Samples in the Training and Validation Studies
Number of Biopsies |
Training Samples |
Validation Samples |
# of Biopsies |
# of Samples |
% |
# of Biopsies |
# of Samples |
% |
1 |
89 |
99% |
1 |
138 |
98% |
2 |
1 |
1% |
2 |
2 |
1% |
3 |
0 |
0% |
3 |
0 |
0% |
4 |
0 |
0% |
4 |
1 |
1% |
Total |
90 |
|
Total |
141 |
|
Table 6: Aggressive Prostate Cancer Autoantibody-Binding Peptides and their Functions
Autoantibody-Binding Peptides |
Function |
ADP-Ribosylation Factor 6 (ARF 6) |
regulates actin cytoskeleton remodeling vesicle shedding by tumor cells |
NK3 homeobox 1 (NKX3-1) |
regulates androgen response genes (BMI1) |
5’-UTR-BMI1 |
androgen response gene |
Centrosomal Protein 164kD (CEP 164) |
responsible for spindle pole integrity at centrosome |
3’- UTR- Ropporin |
responsible for ciliary movement in spermatozoa through dynein regulation |
Aurora Kinase Interacting Protein 1 (AURKAIP-1) |
regulates androgen response genes (TWIST1) |
Casein Kinase 2, alpha prime polypeptide (CSNK2A2) |
regulates cell cycle and cellular division |
hnRNPA1 |
up regulated in PCa |
KDELR1 |
SRC pathway regulator |
SMAD3 |
tumorigenesis |
BRD2 |
transcription regulator |
FKBP4 |
chaperon protein |
Sample collection and processing
2021 Copyright OAT. All rights reserv
Samples and annotated data were obtained from University of Michigan, Johns Hopkins University, and the Michigan Institute of Urology. All samples in the autoantibody-binding peptide discovery study, the training set and the validation study were prostate cancer positive. There were no differences between age and PSA levels across the cohorts. Samples were shipped to a central laboratory were processed or stored upon receipt.
Each subject provided written informed consent, and no patients were compensated for participating. Test results were not provided to the clinical sites for patient care, and the laboratory technicians who performed the antibody-binding peptide tests were blinded to patient names and any patient characteristics. The autoantibody-binding peptide discovery study, the training set study and the validation study were all performed in accordance with the Standards for Reporting of Diagnostic Accuracy criteria.
Laboratory-developed test: Novel serum based multiplexed autoantibody assay development
The debate over PSA-based screening [3] highlights the unmet need for non-invasive prostate cancer biomarkers with greater sensitivity and specificity that are capable of distinguishing aggressive disease from indolent disease. To that end, advanced genomic and proteomic information [6] was employed to develop and refine an assay that can quantify the aggressiveness of prostate cancer (Figure 1). Twenty- one phage-peptides were analyzed to determine the specific phage-peptides known to be associated with an immune system response to aggressive prostate cancer [9]. These phage-peptides encompassed a combination of phage and protein markers, including the original Apifiny® eight autoantibodies.
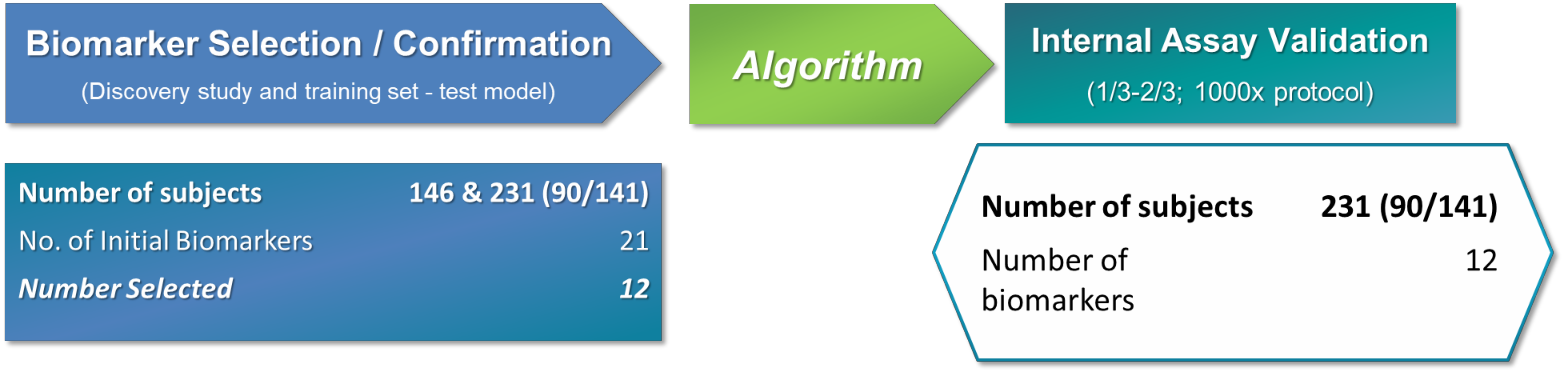
Figure 1. Assay Development and Validation Process
The phage proteins were developed by a technique in which bacteriophages are engineered to fuse a foreign peptide or protein with their capsid proteins and hence expose or display it on their external surface [6]. The immobilized phage may then be used as a screen to see which ligands bind to the expressed fusion protein displayed on the phage surface. The bio-panning process is an iterative process of affinity purification used in the identification and isolation of phages carrying peptides with high affinity and specificity to a given target molecule. Briefly, a library of phage-displayed peptides is incubated with a plate (or bead substrate) coated with the target. Unbound phage particles are washed away, and the bound phages eluted. This process is then repeated several times. This process was used to develop the library of autoantibody-binding peptides used in the biomarker discovery phase of product development [6].
The performance of this expanded serum-based multiplexed autoantibody assay was then tested and validated to assess its ability to discriminate between Gleason 7 or greater from Gleason 6 among patients who had a positive biopsy (Gleason 6 or greater). Using a training set of a total of 231 (90+141) subjects, a unique algorithm was developed to discern between men with low- and high-grade cancers independent of PSA levels. The final algorithm incorporated the twelve autoantibody-binding peptides prioritized for aggressive cancer detection (Table 6).
Statistical analysis
The primary objective was to validate the accuracy of the serum-based multiplexed autoantibody assay to predict high-grade prostate cancer for men aged 40 years or older who had non-aggressive disease and who were not receiving treatment. Thus, only samples which were biopsy-confirmed to be prostate cancer positive were used.
These studies fully complied with the recommendations of the 2012 report from the Institute of Medicine Committee on Omics-Based Predictive Tests [6]. Results of the studies were analyzed by independent biostatisticians.
Building on the understanding that the immune system continuously changes as cancer risk fluctuates, the performance of the peptides was assessed and evaluated as area under the curve (AUC) of the Receiver Operating Characteristic (ROC) curve. The 95% Confidence Intervals (CIs) and comparisons of AUCs were determined using DeLong’s method as implemented in the R package pROC. The combination and predictive value of the multiplexed autoantibody assay was modeled by logistic regression analysis. The assay aimed to identify high-grade (Gleason Score ≥7) prostate cancer.
Pathologically confirmed, low-grade (Gleason Score ≤6) is likely insignificant cancer wherein active surveillance (i.e. no treatment) is the preferred management strategy [5]. The logistic regression model was then applied to estimate the probability of detecting high-grade prostate cancer on biopsy.
Using T7 phage with epitope inserts, autoantibodies against peptides/inserts derived from prostate cancer were identified. At the completion of the autoantibody-binding peptide discovery study, samples were randomly divided into a 1/3 -2/3 protocol whereby approximately one-third of the samples (N=90) were tested, then the remaining (roughly) two-thirds of the samples (N=141) were tested, and this was repeated 1000 times. Raw peptide values were normalized against an internal standard control sample. Logistic regression models were used to model the probability of a sample being high grade cancer as a function of the potential peptides. ROC curves based on fitted multivariate logistic regression models, were also generated to show the relation between sensitivity and specificity for the range of possible cut points.
To determine the optimal number of peptides to include in the final model, we used the following process. For each number of peptides N= 1 to 21 and each normalization method (a total of two including unnormalized) the model with the highest discrimination (based on AUC) with N peptides was calculated. No interactions were considered. The numerically highest cross-validated estimate of AUC was obtained with twelve peptides using the T7 ratio normalization method. To normalize the peptides with T7, for each subject we divided each peptide by that subject’s T7 value. Thus, the final model selected for validation was obtained as the best discriminating model (on the training data) containing twelve peptides using the T7 ratio normalization.
An estimate of predicted risk was defined as the linear combination of the twelve selected peptides each multiplied by their parameter coefficient from the fitted logistic regression model. This risk was calculated for patients in the validation set, and a table of model performance statistics was generated using each risk estimate in the validation data set as a decision threshold for the prediction of high- grade cancer. Specifically, sensitivity, specificity, Positive Predictive Value (PPV) and Negative Predictive Value (NPV) were calculated at each present cut points. In addition, PPV and NPV were calculated using test sensitivity and specificity at 73% study cohort prevalence of aggressive disease. All analysis was performed using R 3.32 (R Core Team, Vienna, Austria).
Results
Selection of the most informative aggressive cancer autoantibody-binding peptides
The performance of individual peptides to predict high-grade prostate cancer was compared. In addition to the AUC, specificity, Negative Predictive Value (NPV), and Positive Predictive Value (PPV) were determined at a fixed sensitivity of approximately 90%. Two cut points were selected. Waterfall Plot of the Autoantibody Assay Scores across the 1/3-2/3 Training/Internal Validation cohort (Figure 2). The lower cut point of 35 was optimized to minimize false negatives. At this level, 12% of patients fell below the cut point, sensitivity was 95% and the false negative rate was only 3.9% for Gleason 7 or greater disease. Positive predictive value was 77%, negative predictive value was 67% and specificity was 28%.
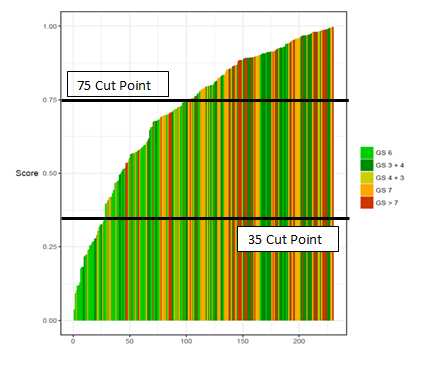
Figure 2. Waterfall Plot of the Autoantibody Assay Score Across the Validation Cohort
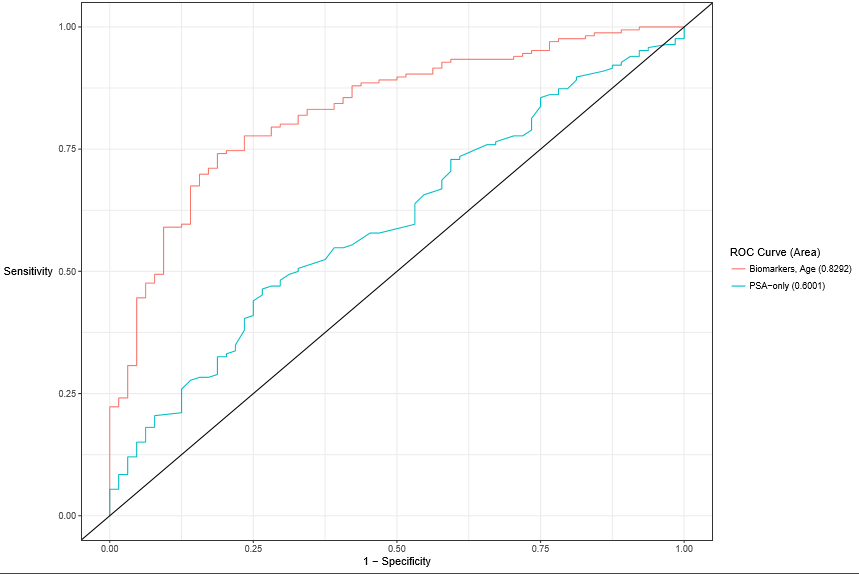
Figure 3: Receiver Operating Curve for the Autoantibody Assay plus Age versus PSA alone.
Clinical validation
The performance characteristics of the assay plus age were evaluated relative to PSA in the independent validation study. Among the 377 men (median age 63.2 years; median PSA 7.4 ng/mL), the expanded autoantibody assay plus age showed significantly better discrimination between patients with Gleason Score 7 or greater from Gleason Score 6 in patients with positive biopsies (AUC 0.83, 95% CI 0.76-0.90) than PSA alone (AUC 0.60, 95% CI 0.52-0.68) (P =0.001). Indeed, a rigorous statistical analysis resulted in two cut points. A test developed with this expanded set of phage-peptide clones detected Gleason 7 or higher at the high cut point of 75 or greater with 71% sensitivity and 84% specificity. Positive predictive value was optimized at 92% and only 4.4% of patients had a false positive result indicating Gleason 6 disease. Negative predictive value was 53%. At the lower cut point of less than 35 with a sensitivity level of 95%, approximately 24.4% of total biopsies could potentially have been avoided, with only 3.9% or six (6) patients with a false negative result (Gleason 7+ disease. Of these six (6) patients whose cancers were missed, a sub-analysis revealed five were Gleason 7 (3+4) and only one was Gleason 7 (4+3). The positive predictive value was 77%, negative predictive value was 67% and specificity was 28%. The validation study population prevalence rate was 73% for aggressive disease. Thus, when compared with standard clinical tools, this expanded multiplexed autoantibody assay provides significant clinical benefit.
Discussion
For the past three decades, serum PSA has been used as a prostate cancer biomarker3. However, due to its inherent limitations, including lack of specificity, PSA screening has proven controversial leading to an intensive search for alternative prostate cancer biomarkers with a better diagnostic and predictive potential [3]. This is especially true in the intermediate range of PSA detection (4-10 ng/mL) [4]. In particular, there is a quest for biomarkers that can distinguish between aggressive and indolent tumors, thereby leading to better treatment decisions.
With the advent of advanced genomic and proteomic technologies, we have in recent years experienced a significant increase in our understanding of prostate cancer biology. As a result, prostate cancer biomarker research has expanded with several US FDA approved and clinical laboratory-based tests that show an improved sensitivity and specificity over PSA. However, unlike the novel autoantibody assay described herein, most recently developed tests measure alternative molecular traits and/or isoforms of the PSA protein. Taken together, measurement of PSA isoforms and dynamics have modestly improved care but are largely hindered by the same issues confounding PSA itself [10].
This study assessed the performance of a novel serum-based multiplexed autoantibody assay using 12 phage-peptide clones plus age compared to PSA alone for separating Gleason Score 7 or greater from Gleason Score 6 in prostate cancer positive patients. The algorithm derived from this model was the best performing assay to detect high-grade prostate cancer on prostate biopsy and was successfully validated in a cohort of 231 (90 + 141) total samples. The assay plus age significantly outperformed PSA alone. Moreover, the algorithm used to generate the performance does not require PSA, its derivatives or any other factors in the assay to be effective.
The assay and the classifier algorithm using the twelve specific biological markers was developed to optimize the positive predictive value at the higher cut point and to minimize false negatives for Gleason 7+ disease at the lower cut point. Thus, when the patient’s score is at or above the high cut point of 75, there is an increased likelihood of high-grade prostate cancer. Scores at or above 75 demonstrated a positive predictive value of 92% for aggressive prostate cancer (Gleason 7+) with only a 4.4% of patients receiving a false positive rate indicating Gleason 6 disease.
When a patient’s score is below the cut point, there is a decreased likelihood of high-grade prostate cancer. Scores below 35 demonstrated a 3.9% false negative rate for aggressive disease (Gleason 7+). Of these six (6) patients whose cancers were missed, a sub-analysis revealed five were Gleason 7 (3+4) and only one was Gleason 7 (4+3).
Consequently, when combined with currently acceptable methods of risk assessment, the results of this test will likely enable objective clinical risk assessment and patient management. For patients on active surveillance, additional insight into the risk of aggressive disease may help prevent multiple biopsies, limiting patient anxiety and the risk of complications including bleeding and infection. As well, supplementary understanding may help provide unique information on the immune system’s response to prostate cancer to monitor progression. Finally, because this novel assay measures specific autoantibodies produced by the immune system in response to the presence of aggressive disease, it may help to minimize the potential for overtreatment of prostate cancer.
The current study has several limitations, including the need to more fully assess the test in other races, as well as to determine how other conditions (such as obesity and its pro-inflammatory state, or steroid use) may affect the assay’s performance. Moreover, the outcome of this study was ≥G7 on biopsy; it is known some G7 disease (especially low volume G7 (3+4)) can be indolent, and some, high-volume G6 can be clinically meaningful. Finally, this study’s outcome was biopsy; it is well documented that approximately 25% of biopsies will return a false-negative result [7]. Ideally, long-term follow-up including prostatectomy data are needed to verify this as a marker of aggressive prostate cancer.
Conclusions
This study evaluated a novel, cancer-specific biomarker assay based on autoantibody signatures that could be used as a noninvasive risk assessment aid for men with non-aggressive disease who are not receiving treatment. When results of this assay are combined with traditional clinical risk factors, risk stratification for high-grade prostate cancer and biopsy decision making are improved compared to current methods in clinical practice. We hypothesize patients with Gleason Score 6 will exhibit the greatest benefits from employing the assay as they may likely avoid both unnecessary prostate biopsy and subsequent overtreatment. We further hypothesize the assay will thus significantly reduce costs to the healthcare system while further improving patient’s quality of care. Providers and their patients diagnosed with indolent prostate cancer may consider using this novel assay prior to proceeding with prostate biopsy or other treatments.
Acknowledgments
We are indebted to Arul Chinnaiyan, M.D., Ph.D. for his continued encouragement of this work, to Dr. Robert Reinert for his guidance, and to Jim Corrigan and the entire Armune Bioscience and MagArray Inc staff for their assistance in the laboratory.
Disclosures
MagArray Inc provided all financial support for the work reported in this article. Drs. Hafron and Freedland are speakers and consultants for Armune, Bioscience.
References
- Klotz L (2017) Active surveillance for low-risk prostate cancer. Curr Opin Urol 27: 225-230. [Crossref]
- Cooperberg MR, Carroll PR (2015) Trends in Management for Patients with Localized Prostate Cancer, 1990-2013. JAMA 314: 80-82. [Crossref]
- Moyer VA (2012) Screening for prostate cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med 157:120-134. [Crossref]
- Schipper M, Wang G, Giles N, Ohrnberger J (2015) Novel prostate cancer biomarkers derived from autoantibody signatures. Transl Oncol 8: 106-111. [Crossref]
- https://www.auanet.org/guidelines/clinically-localized-prostate-cancer-new-
- Wang X, Yu J, Sreekumar A, Varambally S, Shen R, et al. (2005) Autoantibody signatures in prostate cancer. N Engl J Med 353: 1224-1235. [Crossref]
- Wang X, Hafron JM, Freedland SJ, Yu H, Juang A, et. al. (2017) Risk assessment for high-grade prostate cancer using a novel cancer-specific biomarker assay derived from autoantibody signatures. Biomed Res Clin Prac 2: 1-7.
- Freedland S SS, Kapphahn K, Howard L, Ohmberger J, Hafron J (2017) A novel serum based multiplexed 21 autoantibody assay to predict high-grade prostate cancer at initial biopsy. The Journal of Urology 197: 4S
- Micheel CM, Nass SJ, Omenn GS (2012) Evolution of Translational Omics: Lessons Learned and the Path Forward. Washington (DC): National Academies Press (US. [Crossref]
- Prensner JR, Rubin MA, Wei JT, Chinnaiyan AM (2012) Beyond PSA: the next generation of prostate cancer biomarkers. Sci Transl Med 4: 127rv3. [Crossref]



