Abstract
It is well-documented that codon usage biases affect gene translational efficiency; however, it is less known if viruses share their host’s codon usage motifs. We determined that human-infecting viruses share similar codon usage biases as proteins that are expressed in tissues the viruses infect. By performing 7,052,621 pairwise comparisons of genes from humans versus genes from 113 viruses that infect humans, we determined which codon usage motifs were most highly correlated. We found that 16 viruses averaged a significant correlation in codon usage with over 500 human genes per viral gene, 58 viruses were highly correlated with an average of at least 100 human genes per viral gene, and 37 viruses were significantly correlated with an average of at least one human gene per viral gene at an alpha level of 7.09 × 10-9 (0.05 alpha / 7,052,621 comparisons). Only two viruses were not highly correlated with an average of one human gene per viral gene. While relatively few of the interactions were previously documented, the high statistical correlations suggest that researchers may be able to determine which tissues a virus is most likely to infect by analyzing codon usage biases.
Key words
codon usage bias, host, human, virus, virus-host interactions
Introduction
Amino acids are encoded by DNA triplets known as codons; however, since there are only 20 canonical amino acids and 64 possible codons, multiple codons encode a single amino acid [1]. The majority of amino acids are encoded by 2-6 different codons. Despite multiple codons encoding a single amino acid, codon usage is not random in most species [2-5]. Various species, including many plant species, E. coli and Drosophila, also maintain DNA triplet preferences, or codon usage biases, over time in both intronic and exonic regions [6-8].
It is generally accepted that non-random mutations occur more frequently at the third position in the codon, and codon bias persists through selection [9,10]. Numerous biological factors create evolutionary pressure to use certain codons. First, an incomplete set of transfer RNAs (tRNAs) or unequal expression of tRNA anticodons within a tissue or species creates pressure for codons with complementary tRNAs available. Second, translational speed may either increase or decrease depending on the codon used, creating pressure to select codons for which translational efficiency matches the needs of the tissue/cell (i.e. suboptimal codons might be preferential to some species for increased translational efficiency, while in other instances suboptimal codons might decrease translational efficiency) [10,11]. Finally, codon usage bias primarily affects the translation of a gene and is a main determinant of gene expression [12].
Recently, significant correlations for codon usage preferences between RNA viruses (e.g. SBV and KV) and their host, the honeybee, were reported [13]. They proposed that such similarities resulted from co-evolution, which typically occurs in a leapfrog fashion (i.e. as the host evolves to combat the parasite, the parasite evolves to adapt to the new conditions).
We aimed to determine whether the same relationship exists between human and viral genes expressed in tissues targeted by the virus. We analyzed 19,482 human proteins, and compared their codon usage biases against 113 viruses that infect human hosts. We found significant correlations for many viral and human proteins, and where tissue information was available, the top correlated human protein was frequently highly expressed in the tissue type targeted by the virus.
Materials and methods
Data collection and cleaning
We used gene annotations from the General Feature Format (GFF) and GFF3 files from the National Center for Biotechnology Information (NCBI) to extract the reference viral and human sequences [14-16]. Since the reference genome is intended to most accurately represent an average individual in a species, we downloaded all reference sequence data, including the corresponding gene annotations, from NCBI. Similar to the methods used by [17], when multiple isoforms were annotated, the longest isoform was always chosen as the representative isoform for that gene, and we removed all genes with any annotated translational exceptions (e.g., translational, unclassified transcription discrepancy, suspected errors, etc.). These filters had only a minor effect on our data because they eliminated less than 5% of the total sequences. All 19,482 sequence accession numbers can be found in the NCBI database by downloading the complete genome annotations for Homo sapiens; the accession numbers for each virus and their highest correlating genes are located in Table S1.
|
Virus Accession Number
|
Virus Protein Name
|
Pearson’s R Correlation Value
|
P-value
|
Highest Correlating Protein Accession Number
|
Protein Common Name
|
|
NC_000883
|
NS1
|
0.764596741
|
1.94E-13
|
NP_002763.2
|
TMPRSS15
|
|
NC_000898
|
U90
|
0.931483267
|
6.40E-29
|
NP_112561.2
|
TEX15
|
|
NC_001348
|
ICP4
|
0.798569441
|
2.68E-15
|
NP_787081.2
|
FAM181B
|
|
NC_001352
|
E1
|
0.725454272
|
1.20E-11
|
NP_037485.2
|
TMOD4
|
|
NC_001354
|
Pos: 951-2795
|
0.804857764
|
1.11E-15
|
NP_001273387.1
|
USP7
|
|
NC_001355
|
E1
|
0.798328333
|
2.77E-15
|
NP_940841.1
|
KBTBD3
|
|
NC_001356
|
E1
|
0.903438527
|
1.74E-24
|
NP_001138663.1
|
FAM200B
|
|
NC_001357
|
E1
|
0.805278655
|
1.05E-15
|
NP_940841.1
|
KBTBD3
|
|
NC_001405
|
L1
|
0.865302979
|
2.94E-20
|
NP_001073990.2
|
RASSF10
|
|
NC_001430
|
Pos: 727-7311
|
0.837550489
|
6.34E-18
|
NP_000123.1
|
F8
|
|
NC_001436
|
Pr55
|
0.752880597
|
7.22E-13
|
NP_001092872.1
|
CCNK
|
|
NC_001454
|
L3
|
0.792140958
|
6.41E-15
|
NP_612426.1
|
KTI12
|
|
NC_001457
|
Pos: 5345-6895
|
0.859158203
|
1.06E-19
|
NP_061854.1
|
DNAJC10
|
|
NC_001458
|
Pos: 822-2678
|
0.847795937
|
9.88E-19
|
NP_001273176.1
|
RALGPS2
|
|
NC_001460
|
E1B
|
0.806525776
|
8.74E-16
|
NP_001116801.1
|
ZBTB1
|
|
NC_001472
|
Pos: 742-7290
|
0.800822126
|
1.96E-15
|
NP_005224.2
|
EPHA3
|
|
NC_001488
|
Pos: 807-2108
|
0.748225962
|
1.19E-12
|
NP_001073882.3
|
NOBOX
|
|
NC_001490
|
Pos: 629-7168
|
0.891321462
|
5.65E-23
|
NP_002175.2
|
IL6ST
|
|
NC_001526
|
L1
|
0.807134439
|
8.00E-16
|
NP_942089.1
|
MAP4K5
|
|
NC_001531
|
Pos: 961-2781
|
0.852165343
|
4.29E-19
|
NP_079114.3
|
THNSL1
|
|
NC_001576
|
Pos: 791-2836
|
0.785723092
|
1.48E-14
|
NP_899059.1
|
RAB27A
|
|
NC_001583
|
Pos: 878-2794
|
0.787008282
|
1.26E-14
|
NP_940841.1
|
KBTBD3
|
|
NC_001586
|
Pos: 850-2778
|
0.799660538
|
2.31E-15
|
NP_940841.1
|
KBTBD3
|
|
NC_001587
|
Pos: 5430-7016
|
0.749586507
|
1.03E-12
|
NP_057654.2
|
ERGIC2
|
|
NC_001591
|
E1
|
0.845045382
|
1.65E-18
|
NP_078787.2
|
HAUS3
|
|
NC_001593
|
L1
|
0.744112558
|
1.84E-12
|
NP_001167579.1
|
ZBED6
|
|
NC_001595
|
Pos: 5798-7315
|
0.770647823
|
9.56E-14
|
NP_001273644.1
|
AGTPBP1
|
|
NC_001596
|
E1
|
0.844374112
|
1.86E-18
|
NP_940841.1
|
KBTBD3
|
|
NC_001612
|
Pos: 751-7332
|
0.842341207
|
2.70E-18
|
NP_001116105.1
|
CPS1
|
|
NC_001617
|
Pos: 619-7113
|
0.86771873
|
1.74E-20
|
NP_002175.2
|
IL6ST
|
|
NC_001664
|
IE1
|
0.893813269
|
2.86E-23
|
NP_653091.3
|
CASC5
|
|
NC_001676
|
Pos:828-2729
|
0.787967453
|
1.11E-14
|
NP_940841.1
|
KBTBD3
|
|
NC_001690
|
E1
|
0.855417316
|
2.26E-19
|
NP_001092688.1
|
RAD51AP2
|
|
NC_001691
|
E1
|
0.876751214
|
2.23E-21
|
NP_940841.1
|
KBTBD3
|
|
NC_001693
|
E1
|
0.894934035
|
2.10E-23
|
NP_940841.1
|
KBTBD3
|
|
NC_001716
|
IE1
|
0.927833476
|
3.03E-28
|
NP_001073973.2
|
RBM44
|
|
NC_001722
|
Pos: 1103-2668
|
0.737893765
|
3.50E-12
|
NP_002408.3
|
MKI67
|
|
NC_001781
|
L
|
0.876166171
|
2.56E-21
|
NP_065982.1
|
KIAA1586
|
|
NC_001796
|
Pos: 8646-15347
|
0.903986563
|
1.47E-24
|
NP_065982.1
|
KIAA1586
|
|
NC_001798
|
UL39
|
0.904920752
|
1.10E-24
|
NP_036567.2
|
SHC2
|
|
NC_001802
|
Pr55
|
0.78047161
|
2.89E-14
|
NP_001093866.1
|
C2orf73
|
|
NC_001806
|
UL30
|
0.90801467
|
4.15E-25
|
NP_055778.2
|
SBNO2
|
|
NC_001897
|
Pos: 703-7242
|
0.890389641
|
7.26E-23
|
NP_001017975.3
|
HFM1
|
|
NC_001943
|
Pos: 86-4380
|
0.830734096
|
2.04E-17
|
NP_114161.3
|
SPATA16
|
|
NC_002645
|
Pos: 293-12550
|
0.774229507
|
6.22E-14
|
NP_000099.2
|
DLD
|
|
NC_003266
|
L4
|
0.898683268
|
7.19E-24
|
NP_009115.2
|
NISCH
|
|
NC_003443
|
L
|
0.839684044
|
4.35E-18
|
NP_004645.2
|
USP9Y
|
|
NC_003461
|
L
|
0.866879002
|
2.09E-20
|
NP_065982.1
|
KIAA1586
|
|
NC_004104
|
E1
|
0.68207836
|
5.44E-10
|
NP_899059.1
|
RAB27A
|
|
NC_004148
|
L
|
0.867913209
|
1.67E-20
|
NP_065982.1
|
KIAA1586
|
|
NC_004295
|
VP1
|
0.773099678
|
7.13E-14
|
NP_114414.2
|
EIF2A
|
|
NC_004500
|
E1
|
0.880929983
|
8.18E-22
|
NP_004645.2
|
USP9Y
|
|
NC_005134
|
E1
|
0.851299523
|
5.07E-19
|
NP_001138663.1
|
FAM200B
|
|
NC_005147
|
Pos: 21507-22343
|
0.820880135
|
1.01E-16
|
NP_064506.3
|
UGGT2
|
|
NC_005831
|
Pos: 287-20475
|
0.750091303
|
9.77E-13
|
NP_037471.2
|
ALG6
|
|
NC_006273
|
IE1
|
0.87654333
|
2.35E-21
|
NP_055478.2
|
KDM4A
|
|
NC_006577
|
Pos: 22942-27012
|
0.756094354
|
5.07E-13
|
NP_852607.3
|
LRRC70
|
|
NC_007018
|
ORF2
|
0.774104535
|
6.31E-14
|
NP_005112.2
|
MED13
|
|
NC_007026
|
Pos: 828-2486
|
0.704735964
|
8.08E-11
|
NP_001024.1
|
RRM1
|
|
NC_007027
|
Pos: 94-1698
|
0.746908872
|
1.37E-12
|
NP_002717.3
|
PREP
|
|
NC_007455
|
VP1
|
0.768556356
|
1.22E-13
|
NP_803875.2
|
PKHD1L1
|
|
NC_007605
|
BALF5
|
0.934931283
|
1.36E-29
|
NP_620124.1
|
RHOT2
|
|
NC_008188
|
E1
|
0.85042555
|
6.00E-19
|
NP_940841.1
|
KBTBD3
|
|
NC_008189
|
E1
|
0.781785258
|
2.45E-14
|
NP_000305.3
|
PTEN
|
|
NC_009333
|
ORF75
|
0.91780911
|
1.47E-26
|
NP_002891.1
|
RBP3
|
|
NC_009334
|
BALF5
|
0.935906758
|
8.64E-30
|
NP_620124.1
|
RHOT2
|
|
NC_009996
|
Pos: 616-7050
|
0.834124398
|
1.15E-17
|
NP_004939.1
|
DSC1
|
|
NC_010329
|
E1
|
0.908048024
|
4.10E-25
|
NP_940841.1
|
KBTBD3
|
|
NC_010810
|
Pos: 956-7837
|
0.825974666
|
4.46E-17
|
NP_004939.1
|
DSC1
|
|
NC_010956
|
L4
|
0.884411516
|
3.44E-22
|
NP_009115.2
|
NISCH
|
|
NC_011202
|
L1
|
0.825443151
|
4.86E-17
|
NP_787072.2
|
EXOC8
|
|
NC_011203
|
L4
|
0.84556954
|
1.50E-18
|
NP_009115.2
|
NISCH
|
|
NC_011800
|
Pos: 1892-2533
|
0.744847797
|
1.71E-12
|
NP_056526.3
|
GLTSCR1
|
|
NC_012042
|
VP1
|
0.776501461
|
4.72E-14
|
NP_005424.1
|
YES1
|
|
NC_012213
|
E1
|
0.843291298
|
2.27E-18
|
NP_001138663.1
|
FAM200B
|
|
NC_012485
|
E1
|
0.883809966
|
4.00E-22
|
NP_940841.1
|
KBTBD3
|
|
NC_012486
|
E1
|
0.902494945
|
2.32E-24
|
NP_001138663.1
|
FAM200B
|
|
NC_012564
|
VP1
|
0.783191043
|
2.05E-14
|
NP_002899.1
|
REL
|
|
NC_012729
|
NS2
|
0.805392124
|
1.03E-15
|
NP_001073932.1
|
DYNC2H1
|
|
NC_012798
|
Pos: 139-6480
|
0.82589364
|
4.52E-17
|
NP_057190.2
|
SCFD1
|
|
NC_012801
|
Pos: 750-7124
|
0.824196863
|
5.94E-17
|
NP_001191195.1
|
GABRA4
|
|
NC_012802
|
Pos: 748-7128
|
0.834958942
|
9.94E-18
|
NP_001161829.1
|
PLA2G7
|
|
NC_012950
|
Pos: 21445-22281
|
0.818600204
|
1.44E-16
|
NP_064506.3
|
UGGT2
|
|
NC_012959
|
Pos: 22707-24845
|
0.842896843
|
2.44E-18
|
NP_009115.2
|
NISCH
|
|
NC_012986
|
Pos: 719-7831
|
0.755617438
|
5.35E-13
|
NP_004215.2
|
GPR50
|
|
NC_013035
|
E1
|
0.900330229
|
4.44E-24
|
NP_940841.1
|
KBTBD3
|
|
NC_014185
|
E1
|
0.928268261
|
2.53E-28
|
NP_940841.1
|
KBTBD3
|
|
NC_014952
|
E1
|
0.879231256
|
1.24E-21
|
NP_940841.1
|
KBTBD3
|
|
NC_014953
|
E1
|
0.904630266
|
1.21E-24
|
NP_940841.1
|
KBTBD3
|
|
NC_014954
|
E1
|
0.895597619
|
1.74E-23
|
NP_940841.1
|
KBTBD3
|
|
NC_014955
|
E1
|
0.905343727
|
9.67E-25
|
NP_940841.1
|
KBTBD3
|
|
NC_014956
|
E1
|
0.903382032
|
1.77E-24
|
NP_940841.1
|
KBTBD3
|
|
NC_015150
|
Pos: c5026-4790, c4437-2632
|
0.897789054
|
9.32E-24
|
NP_060862.3
|
C4orf21
|
|
NC_015630
|
Pos: 381-1076
|
0.54440122
|
3.32E-06
|
NP_689786.2
|
RASEF
|
|
NC_016157
|
Pos: 817-2640
|
0.919910732
|
6.78E-27
|
NP_940841.1
|
KBTBD3
|
|
NC_017993
|
Pos: 805-2610
|
0.859261423
|
1.04E-19
|
NP_940841.1
|
KBTBD3
|
|
NC_017994
|
E1
|
0.868341489
|
1.52E-20
|
NP_940841.1
|
KBTBD3
|
|
NC_017995
|
Pos: 714-2546
|
0.883104334
|
4.77E-22
|
NP_001138663.1
|
FAM200B
|
|
NC_017996
|
Pos: 717-2534
|
0.881915256
|
6.42E-22
|
NP_940841.1
|
KBTBD3
|
|
NC_017997
|
Pos; 703-2502
|
0.825068761
|
5.16E-17
|
NP_112561.2
|
TEX15
|
|
NC_019023
|
E1
|
0.864842857
|
3.24E-20
|
NP_940841.1
|
KBTBD3
|
|
NC_019843
|
orf1ab
|
0.777846368
|
4.00E-14
|
NP_079265.2
|
PGAP1
|
|
NC_020890
|
large T antigen
|
0.894050364
|
2.68E-23
|
NP_001017975.3
|
HFM1
|
|
NC_021483
|
E1
|
0.858044662
|
1.34E-19
|
NP_001092688.1
|
RAD51AP2
|
|
NC_021568
|
Pos: 279-13433, 13433-21514
|
0.731131014
|
6.89E-12
|
NP_066012.1
|
METTL14
|
|
NC_021928
|
Pos: c5033-4821, c4421-2508
|
0.8986358
|
7.29E-24
|
NP_065982.1
|
KIAA1586
|
|
NC_022095
|
L1
|
0.818922205
|
1.37E-16
|
NP_001273644.1
|
AGTPBP1
|
|
NC_022518
|
Pos: 6451-8550
|
0.801092218
|
1.89E-15
|
NP_001121143.1
|
LIFR
|
|
NC_022892
|
E1
|
0.856537918
|
1.81E-19
|
NP_065982.1
|
KIAA1586
|
|
NC_023874
|
Pos: 161-997
|
0.720321018
|
1.95E-11
|
NP_060146.2
|
GIN1
|
|
NC_023891
|
E1
|
0.88293482
|
4.98E-22
|
NP_940841.1
|
KBTBD3
|
|
NC_023984
|
Pos: 1362-7727
|
0.837884709
|
5.98E-18
|
NP_036434.1
|
LPHN2
|
|
NC_024694
|
Pos: 1 - 1113
|
0.676628221
|
8.40E-10
|
NP_054860.1
|
CNTNAP2
|
Table S1. A comprehensive list of the 113 viruses with their highest correlating protein, accompanied by the Pearson’s r correlation and the respective p-value. Bolded rows were found to be insignificant. Unnamed viral proteins are designated by their position numbers in the following format— Pos: start position-stop position.
Codon usage correlation values
To determine if there was a correlation between human and viral codon usage biases, we performed a Pearson’s r correlation test with discrete codon usage counts by comparing total codon usage counts in human and viral coding sequences (CDS). We used Pearson’s r because it uses a product-moment correlation coefficient that is used to determine the correlation between two variables with different units or different magnitudes [18]. Since gene lengths can vary greatly between genes, and genes do not contain all codons, the assumptions for most statistical tools would not be adequately met using the raw data. Furthermore, the high number of zero codon usage counts in some genes meant that a percentage comparison of codon usages using a traditional t-test was unfeasible, even with a transformation. We chose an implementation of Pearson’s r from the package SciPy in Python version 2.7 because Pearson’s r is robust to variations in sequence sizes as well as zero values. Using Pearson’s r, we graphed a linear regression and calculated the R2 coefficients of determination and p-values by plotting the discrete codon counts from each gene within each virus against each human gene. Next, we ranked the correlation of codon usage between viral and human genes from highest to lowest. We corrected for multiple tests using a Bonferroni correction; the significance threshold used was 7.09 × 10-9 (0.05/7,052,621 total comparisons). We obtained the highest correlations when the viral and human protein codon usage motifs were most similar.
Human tissue comparisons
We determined which proteins were expressed in each human tissue by querying each highly correlated human protein against the Human Protein Atlas [19,20]. We checked the top correlating human proteins for each virus (113 total proteins) to determine in which tissues they were most highly expressed. While many proteins were expressed in low levels throughout the body, we were most concerned with high expression areas, and only the high expression areas were compared in this study.
Results
Of the 113 viruses analyzed, we found that on average, each viral gene in 16 viruses was significantly correlated with more than 500 human proteins (Table S2). Of the remaining 97 viruses, 58 were significantly correlated with at least 100 human proteins per viral gene, and 37 were significantly correlated with at least one human gene per viral gene on average at a p-value <7.09 × 10-9. Only two viruses, Human papillomavirus type 90 (NC_004104) and Human gyrovirus type 1 (NC_015630) were not significantly correlated with the codon usage of at least one human gene per viral gene, on average.
|
Virus Accession Number
|
Virus Name
|
Virus Protein Name
|
Protein Accession Number
|
Protein Name
|
Correlation %
|
P-value
|
|
NC_009334
|
Human herpesvirus 4
|
BALF5
|
NP_620124.1
|
RHOT2
|
93.6
|
8.64E-30
|
|
NC_007605
|
Human herpesvirus 4 (wild type)
|
BALF5
|
NP_620124.1
|
RHOT2
|
93.5
|
1.36E-29
|
|
NC_000898
|
Human herpesvirus 6B
|
U90
|
NP_112561.2
|
TEX15
|
93.1
|
6.40E-29
|
|
NC_014185
|
Human papillomavirus 121
|
E1
|
NP_940841.1
|
KBTBD3
|
92.8
|
2.53E-28
|
|
NC_001716
|
Human herpesvirus 7
|
IE1
|
NP_001073973.2
|
RBM44
|
92.8
|
3.03E-28
|
|
NC_016157
|
Human papillomavirus 126
|
Pos: 817-2640
|
NP_940841.1
|
KBTBD3
|
92.0
|
6.78E-27
|
|
NC_009333
|
Human herpesvirus 8
|
ORF75
|
NP_002891.1
|
RBP3
|
91.8
|
1.47E-26
|
|
NC_010329
|
Human papillomavirus 88
|
E1
|
NP_940841.1
|
KBTBD3
|
90.8
|
4.10E-25
|
|
NC_001806
|
Human herpesvirus 1
|
UL30
|
NP_055778.2
|
SBNO2
|
90.8
|
4.15E-25
|
|
NC_014955
|
Human papillomavirus 132
|
E1
|
NP_940841.1
|
KBTBD3
|
90.5
|
9.67E-25
|
Table 1. Here we report the top-ten codon usage bias correlations (Pearson’s r values) between a virus and a human protein with their respective p-values (all under 10-25), demonstrating that viruses and proteins in their host (humans) share high codon biases. Unnamed viral proteins are designated by their position numbers in the following format— Pos: start position-stop position.
|
Virus Accession Number
|
Number of Genes in Virus
|
Number of Highly Correlating Genes in Humans
|
Number of Highly Correlating Human Proteins per Viral Protein
|
|
NC_015630
|
3
|
0
|
0
|
|
NC_004104
|
7
|
4
|
0.57
|
|
NC_012986
|
1
|
1
|
1
|
|
NC_001436
|
6
|
7
|
1.17
|
|
NC_024694
|
4
|
13
|
3.25
|
|
NC_001488
|
6
|
27
|
4.5
|
|
NC_011800
|
6
|
28
|
4.67
|
|
NC_007026
|
2
|
15
|
7.5
|
|
NC_005831
|
6
|
47
|
7.83
|
|
NC_001722
|
9
|
91
|
10.11
|
|
NC_001352
|
7
|
91
|
13
|
|
NC_023874
|
2
|
32
|
16
|
|
NC_001595
|
6
|
104
|
17.33
|
|
NC_001357
|
8
|
152
|
19
|
|
NC_001454
|
34
|
655
|
19.26
|
|
NC_006577
|
8
|
165
|
20.63
|
|
NC_021568
|
2
|
50
|
25
|
|
NC_001576
|
7
|
221
|
31.57
|
|
NC_001587
|
6
|
219
|
36.5
|
|
NC_001348
|
73
|
2843
|
38.95
|
|
NC_001593
|
7
|
331
|
47.29
|
|
NC_000883
|
6
|
317
|
52.83
|
|
NC_019843
|
11
|
582
|
52.91
|
|
NC_001355
|
9
|
478
|
53.11
|
|
NC_001460
|
36
|
1950
|
54.17
|
|
NC_001583
|
6
|
328
|
54.67
|
|
NC_001676
|
7
|
391
|
55.86
|
|
NC_001526
|
8
|
456
|
57
|
|
NC_008189
|
6
|
353
|
58.83
|
|
NC_001802
|
10
|
629
|
62.9
|
|
NC_002645
|
8
|
613
|
76.63
|
|
NC_001586
|
6
|
517
|
86.17
|
|
NC_015150
|
5
|
435
|
87
|
|
NC_007027
|
1
|
93
|
93
|
|
NC_011202
|
38
|
3637
|
95.71
|
|
NC_007455
|
4
|
392
|
98
|
|
NC_001781
|
11
|
1079
|
98.09
|
|
NC_017997
|
7
|
691
|
98.71
|
|
NC_001354
|
11
|
1096
|
99.64
|
|
NC_012950
|
12
|
1268
|
105.67
|
|
NC_005147
|
9
|
970
|
107.78
|
|
NC_012042
|
4
|
438
|
109.5
|
|
NC_004500
|
7
|
787
|
112.43
|
|
NC_013035
|
7
|
837
|
119.57
|
|
NC_008188
|
6
|
720
|
120
|
|
NC_004295
|
6
|
747
|
124.5
|
|
NC_022095
|
6
|
750
|
125
|
|
NC_012564
|
4
|
555
|
138.75
|
|
NC_004148
|
9
|
1314
|
146
|
|
NC_001405
|
38
|
5628
|
148.11
|
|
NC_000898
|
104
|
15694
|
150.9
|
|
NC_012485
|
7
|
1083
|
154.71
|
|
NC_006273
|
169
|
26217
|
155.13
|
|
NC_001664
|
88
|
13960
|
158.64
|
|
NC_012213
|
5
|
801
|
160.2
|
|
NC_003461
|
10
|
1706
|
170.6
|
|
NC_003266
|
38
|
7275
|
191.45
|
|
NC_001798
|
77
|
14790
|
192.08
|
|
NC_022892
|
6
|
1160
|
193.33
|
|
NC_010956
|
38
|
7500
|
197.37
|
|
NC_017993
|
7
|
1382
|
197.43
|
|
NC_001690
|
7
|
1464
|
209.14
|
|
NC_021483
|
7
|
1467
|
209.57
|
|
NC_001596
|
7
|
1470
|
210
|
|
NC_014953
|
7
|
1498
|
214
|
|
NC_012959
|
36
|
7762
|
215.61
|
|
NC_001591
|
6
|
1327
|
221.17
|
|
NC_014952
|
7
|
1601
|
228.71
|
|
NC_011203
|
39
|
9069
|
232.54
|
|
NC_001531
|
8
|
1903
|
237.88
|
|
NC_012729
|
5
|
1212
|
242.4
|
|
NC_003443
|
7
|
1720
|
245.71
|
|
NC_020890
|
5
|
1235
|
247
|
|
NC_010329
|
7
|
1744
|
249.14
|
|
NC_012486
|
7
|
1768
|
252.57
|
|
NC_001691
|
7
|
1771
|
253
|
|
NC_023891
|
7
|
1843
|
263.29
|
|
NC_001356
|
7
|
1844
|
263.43
|
|
NC_021928
|
7
|
1879
|
268.43
|
|
NC_005134
|
7
|
1893
|
270.43
|
|
NC_014956
|
7
|
1894
|
270.57
|
|
NC_001796
|
8
|
2167
|
270.88
|
|
NC_016157
|
7
|
1969
|
281.29
|
|
NC_001457
|
7
|
1980
|
282.86
|
|
NC_014954
|
7
|
1981
|
283
|
|
NC_014955
|
7
|
2051
|
293
|
|
NC_017994
|
7
|
2061
|
294.43
|
|
NC_014185
|
7
|
2076
|
296.57
|
|
NC_009333
|
86
|
26437
|
307.41
|
|
NC_001458
|
7
|
2182
|
311.71
|
|
NC_001693
|
7
|
2316
|
330.86
|
|
NC_001806
|
77
|
26054
|
338.36
|
|
NC_019023
|
6
|
2070
|
345
|
|
NC_017996
|
7
|
2500
|
357.14
|
|
NC_007018
|
2
|
769
|
384.5
|
|
NC_001716
|
86
|
33651
|
391.29
|
|
NC_017995
|
7
|
2784
|
397.71
|
|
NC_001943
|
2
|
1088
|
544
|
|
NC_022518
|
1
|
592
|
592
|
|
NC_001472
|
1
|
753
|
753
|
|
NC_007605
|
95
|
85227
|
897.13
|
|
NC_009334
|
80
|
82905
|
1036.31
|
|
NC_001612
|
1
|
1133
|
1133
|
|
NC_009996
|
1
|
1157
|
1157
|
|
NC_001617
|
1
|
1193
|
1193
|
|
NC_010810
|
1
|
1223
|
1223
|
|
NC_012802
|
1
|
1408
|
1408
|
|
NC_001490
|
1
|
1423
|
1423
|
|
NC_012798
|
1
|
1437
|
1437
|
|
NC_023984
|
1
|
1453
|
1453
|
|
NC_012801
|
1
|
1482
|
1482
|
|
NC_001430
|
1
|
1720
|
1720
|
|
NC_001897
|
1
|
1918
|
1918
|
|
Average
|
15.74
|
4161.41
|
303.36
|
|
Total
|
1779
|
470239
|
34279.52
|
Table S2. A comprehensive list of the 113 viruses with the number of genes in the virus, the number of highly correlating human genes, and the number of highly correlating human proteins per viral protein. Viruses are ordered in accending order based on the number of highly correlating human genes per viral gene.
The viruses listed in Table 1 have the highest Pearson r correlation values of all comparisons made, with their codon usages strongly correlating to their host codon usages (p-value<10-25). Four of the top 10 correlations in Table 1 belong to the group of 16 viruses that strongly correlate to over 500 human proteins per viral gene on average, and the rest of them belong to the group of 58 with significant correlations with at least 100 human genes significantly correlating to each viral gene, on average. Overall, the average correlation of the 113 viruses with the top hit from each virus was 83.1%, meaning about 83% of the codon usage bias in the virus also existed in the human host protein. Each viral protein strongly correlated to an average of 303 human genes.
To demonstrate the strong correlations in codon usage bias, we plotted codon usage for several representative viral proteins compared to the human protein with the strongest correlation (Figure 1).
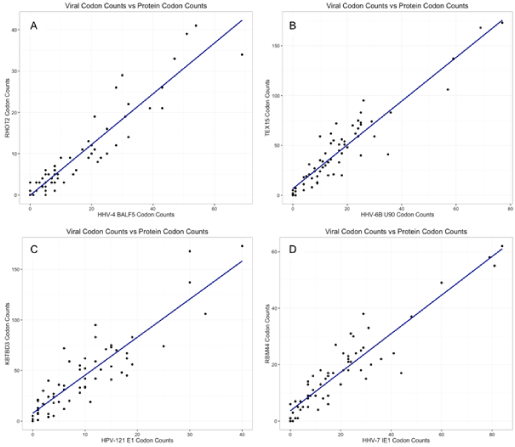
<p><strong>Figure 1. Codon counts. </strong>Four of the highest correlating virus-protein pairs found in Table 1 are displayed. We plotted codon counts for the viral protein (X-axis) against the human protein’s codon counts (Y-axis). Each graph has 64 points, each representing a codon. Points near the top right are used at a higher rate than points near the bottom left. The line represents the result of a best-fit linear model, indicating that there is a strong correlation--as protein codon usage increases, so does the codon usage count of the respective virus. Residual plots of the linear regression were also analyzed and appear to fit the assumptions of the model. (A) displays RHOT2 vs HHV-4 (correlation of 93.6%), (B) shows TEX15 vs HHV-6B (correlation of 93.1%), (C) shows KBTBD3 vs HPV-121 (correlation of 92.8%), and (D) displays RBM44 vs HHV-7 (correlation of 92.8%). See Table 1 for more information on these pairs.</p>
Finally, we analyzed the correlations of codon usage biases for human proteins expressed in tissues infected by a specific virus. With the exception of sexually transmitted diseases (STDs), tissue information was incomplete for many viruses, and further exacerbating this problem is that many human proteins expressed in a specific tissue were also expressed in many other tissues. We report all known tissue information in Table S3, and in Table 2 list representative viruses with their highest correlating protein and affected tissues.
Discussion
The high number of proteins significantly correlated with each virus suggests that humans and human-host viruses share similar codon usage biases. For example, each of the 80 Human herpesvirus 4 (HHV-4, NC_009334) genes significantly correlated with 1 to 10,012 human genes with a median of 8,290 highly correlated human genes and an average of 1,036 highly correlated human genes. HHV-4 was previously identified as having a similar codon usage bias to its host cells [21,22], which may provide insights into the efficient proliferation of HHV-4, since it can more readily utilize host tRNA machinery in the tissue types it infects. Indeed, HHV-4 (commonly known as mononucleosis or “the kissing disease”) is one of the most common viruses known to infect humans, with almost 90% of adults having antibodies suggesting previous HHV-4 infection [22]. Herpesviruses overtake host translational machinery through virion host shutoff (vhs), which limits the expression of host mRNA [23], and through the degradation of host mitochondrial DNA [24], although some herpesvirus strains act differently [25]. Our data suggest that herpesvirus is able to co-opt the translational apparatus of the infected cell by closely matching codon usage biases. The virus is able to use existing tRNAs in the cell, which are not being used by the cell due to vhs.
Furthermore, viruses such as HPV-90 (NC_004104) and Human gyrovirus 1 (NC_015630) with fewer correlating proteins typically occur less frequently in human populations. Although limited data exist for the prevalence of HPV-90 in the general population, in general it presents a very low risk to the general population [26,27]. Human gyrovirus 1, which is identical to the Chicken Anemia Virus, is relatively rare and the effects of the virus still remain largely unknown, although it may affect the apoptosis pathway [28,29].
Human-host viruses appear to target tissues where the correlating human protein also has high expression. Although many viruses analyzed were not clearly annotated as infecting a particular human tissue, the viruses with documented tissue interactions were always highly correlated with a protein that was highly expressed in that tissue. For instance, HPV-128 correlates most with the human protein TIGD4, which is mainly expressed in the genitalia. In addition, other STDs were strongly correlated with proteins that were also mainly expressed in genitalia (Table 2, Table S3). We note that viruses tend to share the same codon usage biases as at least one protein that is highly expressed in the disease targeted area, further emphasizing our conclusion that viral and host codon usage biases are highly correlated.
|
Accession Number
|
Virus Name
|
Virus Protein
|
Correlating Human Protein
|
Protein’s Expression Location
|
|
NC_004500
|
HPV 92
|
E1
|
MSH4
|
Testis
|
|
NC_022095
|
HPV 179
|
L1
|
HLTF
|
Testis
|
|
NC_014952
|
HPV 128
|
E1
|
TIGD4
|
Testis, vagina
|
|
NC_001691
|
HPV 50
|
E1
|
TEX15
|
Testis
|
|
NC_001405
|
HPV 18
|
L1
|
MRC2
|
Soft tissue, testis, endometrium
|
|
NC_001354
|
HPV 41
|
USP7
|
SLC12A2
|
Digestive tract, breast, placenta
|
|
NC_000898
|
HHV 6
|
U90
|
ELTD1
|
Gallbladder, breast, smooth muscle
|
|
NC_019023
|
HPV 166
|
E1
|
OTOGL
|
Cervix, testis
|
|
NC_009334
|
HHV 4
|
BALF5
|
SPTB
|
Epididymis
|
|
NC_010329
|
HPV 88
|
E1
|
RAD51AP2
|
Seminal Vesicle, Fallopian Tube
|
|
NC_004500
|
HPV 92
|
E1
|
USP9Y
|
Prostate
|
Table 2. A selection of viral proteins and their top correlating human proteins, along with the human protein’s documented area of expression. These results show that viral codon usage biases highly correlate with the codon usage biases of human proteins that are found within tissues that the viruses are known to promote symptomatic issues.
|
Virus Accession Number
|
Highest Correlating Human Protein Accession Number
|
Region(s) Where Human Protein is Most Highly Expressed
|
|
NC_000883
|
NP_002763.2
|
Stomach glandular cells
|
|
NC_000898
|
NP_112561.2
|
Testis, urinary tract, and brain
|
|
NC_001348
|
NP_787081.2
|
Myocytes in heart muscle, lateral ventricle, cerebral cortex,
|
|
|
|
hippocampus
|
|
NC_001352
|
NP_037485.2
|
Myocytes in skeletal muscle, and glandular cells in the stomach.
|
|
NC_001354
|
NP_001273387.1
|
Liver, pancreas, digestive tract, male reproductive system, endocrine
|
|
NC_001355
|
NP_940841.1
|
Skeletal muscle, smooth muscle, epidermal cells, hepatocytes in liver
|
|
NC_001356
|
NP_001138663.1
|
GI-tract, gallbladder, and the blood and immune system
|
|
NC_001357
|
NP_940841.1
|
Smooth muscle cells
|
|
NC_001405
|
NP_001073990.2
|
Stomach, kidney, fallopian tube,
|
|
NC_001430
|
NP_000123.1
|
Adipocytes of soft tissue, placenta, tubule cells in the kidney
|
|
NC_001436
|
NP_001092872.1
|
Hematopoietic cells in bone marrow, glandular cells in the stomach
|
|
NC_001454
|
NP_612426.1
|
Glandular cells of the GI tract, urinary tract cells, adrenal glands
|
|
NC_001457
|
NP_061854.1
|
Glandular cells of the epididymis and the endometrium
|
|
NC_001458
|
NP_001273176.1
|
Testis.
|
|
NC_001460
|
NP_001116801.1
|
Kidney, testis, stomach, esophagus, vagina, skin, lung, and heart
|
|
NC_001472
|
NP_005224.2
|
Low expression everywhere
|
|
NC_001488
|
NP_001073882.3
|
No information found
|
|
NC_001490
|
NP_002175.2
|
Stomach cells, prostate, kidney, liver, pancreas, heart muscle
|
|
NC_001526
|
NP_942089.1
|
Female reproductive system
|
|
NC_001531
|
NP_079114.3
|
Stomach
|
|
NC_001576
|
NP_899059.1
|
Stomach and rectum
|
|
NC_001583
|
NP_940841.1
|
Smooth muscle cells
|
|
NC_001586
|
NP_940841.1
|
Smooth muscle cells
|
|
NC_001587
|
NP_057654.2
|
Heart muscle cells, and some GI-tract cells.
|
|
NC_001591
|
NP_078787.2
|
Stomach
|
|
NC_001593
|
NP_001167579.1
|
GI-tract and female reproductive system
|
|
NC_001595
|
NP_001273644.1
|
Testis
|
|
NC_001596
|
NP_940841.1
|
Smooth muscle cells
|
|
NC_001612
|
NP_001116105.1
|
Stomach and liver
|
|
NC_001617
|
NP_002175.2
|
Stomach cells, prostate, kidney, liver, pancreas, heart muscle
|
|
NC_001664
|
NP_653091.3
|
Testis
|
|
NC_001676
|
NP_940841.1
|
Smooth muscle cells
|
|
NC_001690
|
NP_001092688.1
|
Male reproductive system
|
|
NC_001691
|
NP_940841.1
|
Smooth muscle cells
|
|
NC_001693
|
NP_940841.1
|
Smooth muscle cells
|
|
NC_001716
|
NP_001073973.2
|
Testis
|
|
NC_001722
|
NP_002408.3
|
Blood, immune system
|
|
NC_001781
|
NP_065982.1
|
Seminal vesicle in men, and the breast in women
|
|
NC_001796
|
NP_065982.1
|
Seminal vesicle in men, and the breast in women
|
|
NC_001798
|
NP_036567.2
|
Varied expression everywhere
|
|
NC_001802
|
NP_001093866.1
|
Male reproductive system and GI-tract
|
|
NC_001806
|
NP_055778.2
|
Liver cells, skeletal muscle, cerebral cortex, endocrine glands, lung
|
|
NC_001897
|
NP_001017975.3
|
Lung cells and skeletal muscles
|
|
NC_001943
|
NP_114161.3
|
Testis and cerebellum
|
|
NC_002645 NC_003266
|
NP_000099.2 NP_009115.2
|
Nearly everywhere, except skin
|
| |
|
Skin, gallbladder, cerebellum, heart muscle, adrenal gland, bronchus
|
|
NC_003443
|
NP_004645.2
|
Prostate
|
|
NC_003461
|
NP_065982.1
|
Seminal vesicle in men, and the breast in women
|
|
NC_004104
|
NP_899059.1
|
Stomach and rectum
|
|
NC_004148
|
NP_065982.1
|
Seminal vesicle in men, and the breast in women
|
|
NC_004295
|
NP_114414.2
|
Skin
|
|
NC_004500
|
NP_004645.2
|
Prostate
|
|
NC_005134
|
NP_001138663.1
|
GI-tract, gallbladder, and the blood and immune system
|
|
NC_005147
|
NP_064506.3
|
Testis and the brain
|
|
NC_005831
|
NP_037471.2
|
Both male and female reproductive systems
|
|
NC_006273
|
NP_055478.2
|
Stomach, testis, and brain
|
|
NC_006577
|
NP_852607.3
|
Hippocampus, heart muscle, parathyroid gland
|
|
NC_007018
|
NP_005112.2
|
Bone marrow, and testis
|
|
NC_007026
|
NP_001024.1
|
Testis, lymph nodes, and lateral ventricles
|
|
NC_007027
|
NP_002717.3
|
GI-tract, and endometrium in women
|
|
NC_007455
|
NP_803875.2
|
Spleen and bone marrow
|
|
NC_007605
|
NP_620124.1
|
Stomach, placenta, skeletal muscle, and cerebral cortex
|
|
NC_008188
|
NP_940841.1
|
Smooth muscle cells
|
|
NC_008189
|
NP_000305.3
|
Cerebral cortex
|
|
NC_009333
|
NP_002891.1
|
No information found
|
|
NC_009334
|
NP_620124.1
|
Stomach, placenta, skeletal muscle, and cerebral cortex
|
|
NC_009996
|
NP_004939.1
|
highest expression in the skin keratinocytes
|
|
NC_010329
|
NP_940841.1
|
Smooth muscle cells
|
|
NC_010810
|
NP_004939.1
|
Skin keratinocytes
|
|
NC_010956
|
NP_009115.2
|
Skin, gallbladder, cerebellum, heart muscle, adrenal gland, bronchus
|
|
NC_011202
|
NP_787072.2
|
Adrenal gland, cerebellum, stomach, and placenta
|
|
NC_011203
|
NP_009115.2
|
Skin, gallbladder, cerebellum, heart muscle, adrenal gland, bronchus
|
|
NC_011800
|
NP_056526.3
|
Medium/high expression everywhere
|
|
NC_012042
|
NP_005424.1
|
Testis, stomach, and placenta
|
|
NC_012213
|
NP_001138663.1
|
GI-tract, gallbladder, blood and immune system
|
|
NC_012485
|
NP_940841.1
|
Smooth muscle cells
|
|
NC_012486
|
NP_001138663.1
|
GI-tract, gallbladder, blood and immune system
|
|
NC_012564
|
NP_002899.1
|
Blood, immune system, women reproductive system, and GI-tract
|
|
NC_012729
|
NP_001073932.1
|
GI-tract
|
|
NC_012798
|
NP_057190.2
|
Pancreas, testis, kidney, and placenta
|
|
NC_012801
|
NP_001191195.1
|
Cerebral cortex
|
|
NC_012802
|
NP_001161829.1
|
Appendix, prostate, placenta, lymph node, and spleen
|
|
NC_012950
|
NP_064506.3
|
Testis and the brain
|
|
NC_012959
|
NP_009115.2
|
Skin, gallbladder, cerebellum, heart muscle, adrenal gland, bronchus
|
|
NC_012986
|
NP_004215.2
|
Kidney and smooth muscle tissue
|
|
NC_013035
|
NP_940841.1
|
Smooth muscle cells
|
|
NC_014185
|
NP_940841.1
|
Smooth muscle cells
|
|
NC_014952
|
NP_940841.1
|
Smooth muscle cells
|
|
NC_014953
|
NP_940841.1
|
Smooth muscle cells
|
|
NC_014954
|
NP_940841.1
|
Smooth muscle cells
|
|
NC_014955
|
NP_940841.1
|
Smooth muscle cells
|
|
NC_014956
|
NP_940841.1
|
Smooth muscle cells
|
|
NC_015150
|
NP_060862.3
|
No information available
|
|
NC_015630
|
NP_689786.2
|
GI-tract and urinary tract
|
|
NC_016157
|
NP_940841.1
|
Smooth muscle cells
|
|
NC_017993
|
NP_940841.1
|
Smooth muscle cells
|
|
NC_017994
|
NP_940841.1
|
Smooth muscle cells
|
|
NC_017995
|
NP_001138663.1
|
GI-tract, gallbladder, blood and immune system
|
|
NC_017996
|
NP_940841.1
|
Smooth muscle cells
|
|
NC_017997
|
NP_112561.2
|
Low expression everywhere
|
|
NC_019023
|
NP_940841.1
|
Smooth muscle cells
|
|
NC_019843
|
NP_079265.2
|
Testis, placenta and parathyroid gland
|
|
NC_020890
|
NP_001017975.3
|
Lung cells and skeletal muscles
|
|
NC_021483
|
NP_001092688.1
|
Stomach, male reproductive system, and skin
|
|
NC_021568 NC_021928
|
NP_066012.1 NP_065982.1
|
Testis and stomach
|
| |
|
Seminal vesicle in men, and the breast in women
|
|
NC_022095
|
NP_001273644.1
|
Testis
|
|
NC_022518
|
NP_001121143.1
|
Male reproductive tissue and in the heart
|
|
NC_022892
|
NP_065982.1
|
Seminal vesicle in men, and the breast in women
|
|
NC_023874
|
NP_060146.2
|
Tonsil, stomach, and pancreas
|
|
NC_023891
|
NP_940841.1
|
Smooth muscle cells
|
|
NC_023984
|
NP_036434.1
|
Skeletal and smooth muscle, tonsils, small intestine, colon
|
|
NC_024694
|
NP_054860.1
|
Cerebral cortex
|
Table S3. A comprehensive list of where the highest correlating human protein with respect to a human-infecting virus is most highly expressed.
2021 Copyright OAT. All rights reserv
Highly expressed genes have codon biases that utilize highly abundant tRNAs in order for optimal translational and transcriptional speed [12,13,30-33]. The Human Adenovirus E (NP_009115.2), which causes respiratory illness, has an 89.9% codon usage correlation with the NISCH gene, which is mainly expressed in the bronchus. Since NISCH is highly expressed in the tissues that the adenovirus normally infects, the virus is able to take advantage of its codon usage bias similarities with the host proteins to rapidly proliferate and infect additional hosts.
There are other possibilities for the observed shared codon usage biases. For example, co-evolution may have contributed to the appearance of such strong codon bias correlations, in which the host and the virus evolve at similar rates in order to either combat or maintain parasitic infection [34]. Since viruses have smaller genomes, they can selectively evolve more rapidly toward being similar to a preferred host.
While co-evolution and the abundance of optimal tRNAs are thought to allow greater viral spread, determining the exact cause of this correlation remains unexplored. Our extensive analysis of codon usage determined that a strong correlation in codon usage bias exists between human-host viruses and proteins expressed in the human tissues that they infect. Future research should focus on the causes of these correlations.
Authorship and contributorship
JM and PR conceived the idea. JM oversaw all aspects of the project. AH developed the comparison algorithms and ran the comparisons. CM and SW conducted literature searches and wrote sections of the paper. JM and PR were primarily responsible for editing the manuscript. PR mentored the project.
Acknowledgements
We also appreciate Mark Ebbert and Samantha Jensen who provided expert suggestions for the project flow and design.
Funding information
We appreciate the contributions of Brigham Young University and the Fulton Supercomputing Laboratory in supporting our research.
Competing interests
The authors declare that they have no competing interests.
Availability of data and material
All data are freely available from the NCBI database at ftp://ftp.ncbi.nlm.nih.gov/
References
- Crick FH (1968) The origin of the genetic code. J Mol Biol 38: 367-379. [Crossref]
- Ikemura T (1985) Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol 2: 13-34. [Crossref]
- Sharp PM, Li WH (1986) An evolutionary perspective on synonymous codon usage in unicellular organisms. J Mol Evol 24: 28-38. [Crossref]
- Gutman GA, Hatfield GW (1989) Nonrandom utilization of codon pairs in Escherichia coli. Proc Natl Acad Sci U S A 86: 3699-3703. [Crossref]
- Zhang YM, Shao ZQ, Yang LT, Sun XQ, Mao YF, et al. (2013) Non-random arrangement of synonymous codons in archaea coding sequences. Genomics 101: 362-367. [Crossref]
- Akashi H, Goel P, John A (2007) Ancestral inference and the study of codon bias evolution: implications for molecular evolutionary analyses of the Drosophila melanogaster subgroup. PLoS One 2: e1065. [Crossref]
- Yang Z, Nielsen R (2008) Mutation-selection models of codon substitution and their use to estimate selective strengths on codon usage. Mol Biol Evol 25: 568-579. [Crossref]
- Xu W, Xing T, Zhao M, Yin X, Xia G, et al. (2015) Synonymous codon usage bias in plant mitochondrial genes is associated with intron number and mirrors species evolution. PLoS One 10: e0131508.
- Hershberg R, Petrov DA (2008) Selection on codon bias. Annu Rev Genet 42: 287-299. [Crossref]
- Quax TE, Claassens NJ, Söll D, van der Oost J (2015) Codon Bias as a Means to Fine-Tune Gene Expression. Mol Cell 59: 149-161. [Crossref]
- Xu Y, Ma P, Shah P, Rokas A, Liu Y, et al. (2013) Non-optimal codon usage is a mechanism to achieve circadian clock conditionality. Nature 495: 116-120. [Crossref]
- Zhou Z, Dang Y, Zhou M, Li L, Yu CH, et al. (2016) Codon usage is an important determinant of gene expression levels largely through its effects on transcription. Proc Natl Acad Sci U S A 113: E6117-6117E6125. [Crossref]
- Chantawannakul P, Cutler RW (2008) Convergent host-parasite codon usage between honeybee and bee associated viral genomes. J Invertebr Pathol 98: 206-210. [Crossref]
- Pruitt KD, Brown GR, Hiatt SM, Thibaud-Nissen F, Astashyn A, et al. (2014) RefSeq: an update on mammalian reference sequences. Nucleic Acids Res 42: D756-D763. [Crossref]
- Tatusova T, Ciufo S, Fedorov B, O'Neill K, Tolstoy I (2014) RefSeq microbial genomes database: new representation and annotation strategy. Nucleic Acids Res 42: D553-D559. [Crossref]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, et al. (2007) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 35: D5-D12. [Crossref]
- Camiolo S, Melito S, Porceddu A (2015) New insights into the interplay between codon bias determinants in plants. DNA Res 22: 461-470. [Crossref]
- Häne BG, Jäger K, Drexler HG (1993) The Pearson product-moment correlation coefficient is better suited for identification of DNA fingerprint profiles than band matching algorithms. Electrophoresis 14: 967-972. [Crossref]
- Uhlén M, Björling E, Agaton C, Szigyarto CA, Amini B, et al. (2005) A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics 4: 1920-1932. [Crossref]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, et al. (2015) Proteomics. Tissue-based map of the human proteome. Science, 347, 1260419. [Crossref]
- Roychoudhury S, Mukherjee D (2010) A detailed comparative analysis on the overall codon usage pattern in herpesviruses. Virus Res 148: 31-43. [Crossref]
- Virgin HW, Wherry EJ, Ahmed R (2009) Redefining chronic viral infection. Cell 138: 30-50. [Crossref]
- Smiley JR (2004) Herpes simplex virus virion host shutoff protein: immune evasion mediated by a viral RNase? J Virol 78: 1063-1068. [Crossref]
- Saffran HA, Pare JM, Corcoran JA, Weller SK, Smiley JR (2007) Herpes simplex virus eliminates host mitochondrial DNA. EMBO Rep 8: 188-193. [Crossref]
- Duguay BA, Saffran HA, Ponomarev A, Duley SA, Eaton HE, et al. (2014) Elimination of mitochondrial DNA is not required for herpes simplex virus 1 replication. J Virol 88: 2967-2976. [Crossref]
- Schmitt M, Depuydt C, Benoy I, Bogers J, Antoine J, et al. (2013) Prevalence and viral load of 51 genital human papillomavirus types and three subtypes. Int J Cancer 132: 2395-2403. [Crossref]
- Quiroga-Garza G, Zhou H, Mody DR, Schwartz MR, Ge Y (2013) Unexpected high prevalence of HPV 90 infection in an underserved population: is it really a low-risk genotype? Arch Pathol Lab Med 137: 1569-1573. [Crossref]
- Sauvage V, Cheval J, Foulongne V, Gouilh MA, Pariente K, et al. (2011) Identification of the first human gyrovirus, a virus related to chicken anemia virus. J Virol 85: 7948-7950. [Crossref]
- Chaabane W, Cieślar-Pobuda A, El-Gazzah M, Jain MV, Rzeszowska-Wolny J, et al. (2014) Human-gyrovirus-Apoptin triggers mitochondrial death pathway--Nur77 is required for apoptosis triggering. Neoplasia 16: 679-693. [Crossref]
- Grosjean H, Fiers W (1982) Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene 18: 199-209. [Crossref]
- Morton BR (1998) Selection on the codon bias of chloroplast and cyanelle genes in different plant and algal lineages. J Mol Evol 46: 449-459. [Crossref]
- Morton BR, So BG (2000) Codon usage in plastid genes is correlated with context, position within the gene, and amino acid content. J Mol Evol 50: 184-193. [Crossref]
- Merkl R (2003) A survey of codon and amino acid frequency bias in microbial genomes focusing on translational efficiency. J Mol Evol 57: 453-466. [Crossref]
- Parrish CR, Holmes EC, Morens DM, Park EC, Burke DS, et al. (2008) Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol Mol Biol Rev 72: 457-470. [Crossref]

