Abstract
Olfactory Receptors (ORs) are a large family of G protein-coupled receptors predominantly expressed by the main olfactory epithelium at nasal level and are responsible for the generation of smelling sense. Microarray and deep sequencing analyses, however, have demonstrated that ORs are ectopically expressed in various human tissues including testis, kidneys, adipose tissue and liver and their biological functions become to be unrevealed. Molecular and pharmacological approaches have shown that some of these ORs modulate glucose and lipid metabolism at multiple interfaces, suggesting that ORs might be part of the large family of nutrient sensors, i.e. molecular/ cellular machines that respond to a specific nutrient component. By using nutrient-derived agonists it has been shown that ORs effectively modulates glucose and lipid metabolism raising interest on their possible therapeutic application in the treatment of metabolic disorders including dyslipidemia, obesity and metabolic syndrome.
Keywords
Olfactory receptors, glucose metabolism, type 2 diabetes mellitus, lipid metabolism, NAFLD, metabolic syndrome
Abbreviations
ACC: Acetyl-CoA carboxylase; ACIII: Adenylyl cyclase type III; AMPK: AMP-activated protein kinase; ASM: Airway smooth muscle; AzA: Azelaic acid; BAT: Brown adipose tissue; CAC channel: Calcium activated chloride channel; CaMKIV: Calcium /calmodulin-dependent protein kinase IV; cAMP: Cycling adenosine monophosphate; CNG channel: Cyclic nucleotide gated channel; CNVR: Copy number variation region; CREBP: cAMP response element binding protein; CVDs: Cardiovascular diseases; DAG: 1,2-diacylglicerol; ER: Endoplasmic reticulum; FAO: Fatty acids oxidation; FAS: Fatty acid synthase; G protein: GTP-binding protein; G6PC: G6PC; GDP: Guanosine diphosphate; GF: Growth factor; GI: Gastrointestinal; GK: Glucokinase; GLP-1: Glucagon-like peptide 1; GLUT: Facilitated diffusion glucose transporter; GPCR: G protein-coupled receptor; GSIS: Glucose-stimulated insulin secretion; GTP: Guanosine triphosphate; GWAS: Genome-wide association study; HES-1: Hairy and enhancer of split-1; HFD: High fat diet; HGP: Hepatic glucose production; HSL: Hormone sensitive lipase; IP3: Inositol 1,4,5-triphosphate; IP3R: Inositol 1,4,5-triphosphate receptor; IR: Insulin resistance; LXRα: Liver-X-receptor α; MCFA: Medium-chain fatty acid; MetS: Metabolic Syndrome; MOE: Main olfactory epithelium; mtGPAT: Mitochondrial glycerol-3-phosphate acyltransferase; NAFLD: Nonalcoholic fatty liver disease; NASH: Nonalcoholic steatohepatitis; OLFR: Olfactory receptor (murine); OR: Olfactory receptor (human); OSN: Olfactory sensory neuron; PCK1: Phospho-enolpyruvate carboxykinase; PGC-1α: Peroxisome proliferator-activated receptor γ coactivator 1α; PIP2: Phosphatidylinositol 4,5-biphosphate; PKA: Protein kinase A; PLC: Phospholipase C; PPAR-α: Peroxisome proliferator-activated receptor-α; PPAR-γ: Peroxisome proliferator-activated receptor-γ; RF: Risk factor; SCD1: Stearoyl-CoA desaturase 1; SCFA: Short-chain fatty acid; SGLT: Sodium-glucose transporter; SREBP-1a/1c: Sterol regulatory element-binding protein 1a/1c; T2DM: Type 2 diabetes mellitus; TCA: Tricarboxylic acid; TM: Transmembrane; UCP1: Uncoupling protein 1.
Olfactory Receptors (ORs)
Olfactory receptors (ORs) were discovered in 1991 on olfactory cilia of rats’ main olfactory epithelium (MOE) by Richard Axel and Linda B. Buck [1], while investigating mechanisms involved in smelling sense generation [2]. ORs are G protein-coupled receptors (GPCRs) [3] that share the pro-typical architecture of these receptors organized in (i) seven transmembrane α-helices (TM1 to TM7) linked one to another by (ii) three extracellular and three intracellular loops, (iii) an extracellular N-terminus domain responsible for ligand binding and (iv) an intracellular C-terminus domain (generally organized in an additional eighth α-helix) interacting with a heterotrimeric complex (α, ß and γ subunits) of GTP-binding proteins (G proteins) [4] (Figure 1). According to a sequence homology categorization, ORs belong to one of the five sub-families of the class A rhodopsin like GPCR family, which encodes for almost the 90% of all GPCRs [5].
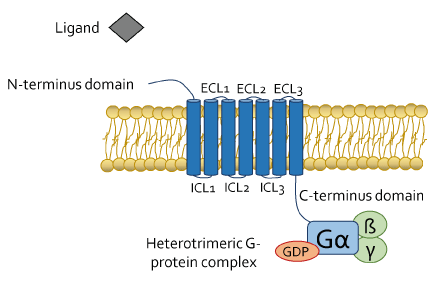
Figure 1. GPCRs structure. The seven transmembrane α-helices counterclockwise arranged are linked one to the other by three extracellular (ECL1-3) and three intracellular (ICL1-3) loops. The extracellular N-terminus domain is deputy to ligand binding, while the intracellular C-terminus domain relates to the heterotrimeric αβγ G-protein complex when the receptor is in its conformational inactive state (unbound ligand) and Gα subunit binds guanosine diphosphate (GDP)
At nasal level, multiple subpopulations of Olfactory Sensory Neurons (OSNs) have been identified, each one characterized by the expression of a single OR protein and, thus, able to respond to different chemosensory stimuli (i.e. different ligands, also known as odorants) [6]. Following the binding to its ligand, the OR becomes activated and promotes the classical G protein signaling pathway, triggering the Adenylyl Cyclase type III (ACIII) activation that is followed by a change in cyclic adenosine monophosphate (cAMP) levels. cAMP induce cyclic nucleotide gated (CNG) channels opening and increase of intracellular Ca++ levels which further stimulates the opening of calcium activated chloride (CAC) channels. This promotes the cell depolarization, action potential generation and neuronal impulse transmission to the central nervous system, allowing the generation of smell perception [7].
ORs are encoded by large family of genes, approximately 1000 genes in the murine genome and at least 400 genes in the human genome and can be classified as class I and class II ORs, depending on their ability to bind and be activated by water-soluble/moderately hydrophobic or highly hydrophobic/volatile compounds, respectively [8].
In addition to the nasal level, an ectopic expression of ORs has been detected in various mammalian tissues [9] including: (i) the testis, where ORs guide spermatogenesis and sperm chemotaxis [10]; (ii) the heart, where ORs seem to be involved in cardiac morphogenesis [11]; (iii) the placenta [12]; (iv) the juxtaglomerular renal apparatus, Olfr78 recognizes and binds the short-chain fatty acid (SCFA) propionate and regulates renin secretion [13,14]; (v) enterochromaffin cells, OR73, OR17-7/11, OR1G1 and OR17-210 promote secretion of serotonin – a tryptophan-derived neurotrasnmitter involved in gut motility and mucus secretion. These later receptors are timulated by terpenoids such as eugenol, thymol and burgeonal [15]. Moreover, ORs have been detected in other organs such as pancreas, adipose tissue and liver [16], where their function seems to be mostly related to regulation of glucose and lipid metabolism [17] (Table 1).
Tissue and/or cell line |
Species |
Receptor(s) |
Main function(s) |
Testis
|
Human
|
OR7A5, OR4D1, OR1D1, hOR17-4, hOR17-2
|
Sperm chemotaxis and chemokinesis [10], [18], [19] |
|
Mouse |
MOR23, MOR244-3, MOR139-3, MOR248-11, MOR31-2, MOR171-31, MOR264-10, MOR256-25, MOR144-1, MOR171-31, MOR13-4, MOR281-1, MOR127-2, MOR13-6, MOR174-6, MOR31-2, MOR248-11 |
Regulation of sperm flagellar motility, chemotaxis, spermatogenesis [20], [21] |
Tongue |
Human |
HTPCR06, HGMP071, JCG1, JCG2, JCG3, JCG4, JCG5, JCG6, JCG9, JCG10, TPCR85, TPCR120 |
Involvement in taste perception [22], [23] |
Heart |
Rat
Human
Mouse |
OL1
OR5E1
MOL2.3 |
Involved in cardiac morphogenesis [11]
Regulation of cardiac function [24]
Not yet defined [25] |
Spleen |
Mouse |
OL-2 |
Not yet defined [26] |
MIN6 |
Mouse |
OL-2 |
Possible regulation of insulin secretion [26] |
Pancreas |
Mouse |
OLFR15, OLFR821 |
Glucose-stimulated insulin secretion (GSIS) [27], [28] |
Blood |
Human |
HPFH1OR |
Not yet defined [29] |
Prostate |
Human |
PSGR |
Overexpression in tumor specimens: potential prostate cancer marker [30], [31] |
Muscle |
Mouse |
MOR23 (also known as OLFR16) |
Involved in muscle regeneration, cell adhesion and migration [32] |
Liver |
Rat
|
PSGR |
Not yet defined [33] |
|
Mouse
Human |
OLFR99, OLFR267, OLFR1393, OLFR1366, OLFR691, OLFR558, OLFR57, OLFR646, OLFR78, OLFR15, OLFR177
OLFR544, OLFR43, OLFR16
OLFR734
OR10J5, OR1A1 |
Involvement in hepatic physiology [16]
Regulation of lipid metabolism (see below)
Gluconeogenesis induction [34]
Regulation of lipid metabolism (see below) |
HepG2 |
Human |
OR1A1, OR10J5 |
Involved in triglycerides synthesis [35], [36] |
Huh7 |
Human |
OR1A2 |
Inhibition of cell proliferation [37] |
3T3-L1 |
Mouse |
OLFR544 |
Triglycerides hydrolyzation [38] |
Adipose tissue |
Mouse
Rat
Human |
OLFR544, OLFR16
OLFR788
OLR984
OR6C3, OR4C11, OR4C6, OR4P4, OR4S2 |
Antiobesogenic effect [36], [38] and brown adipose tissue (BAT) thermogenesis [39]
Antiobesogenic effect [165]
Antiobesogenic effect [165]
Antiobesogenic effect [165] - [167] |
Kidney |
Mouse
Human |
OLFR78
OLFR90, OLFR 1373, OLFR1372
OLFR1393
OLFR31, OLFR99, OLFR545, OLFR691, OLFR693, OLFR1426
OR2T1 |
Involved in renin secretion and blood pression regulation [14]
Not yet defined [13]
Glucose reabsorption [40]
Not yet defined [41]
Not yet identified [41] |
GI tract |
Mouse
Human
|
PSGR
OLFR544, OLFR43
OR73, hOR17-7/11, OR1G1, hOR17-210
OR51E1, OR1A1 |
Not yet defined [33]
Glucose metabolism (GLP-1 and glucagon secretion) [42]
Involved in serotonin secretion [15]
Glucose metabolism (GLP-1 and glucagon secretion) [43] |
Placenta |
Rat
Human |
MOR125-1, MOR126-1, MOR140-1, MOR145-5, MOR216-1, MOR263-9
OR2C1, OR4D6, OR2M7, OR10A6,
OR4F3, OR8A1, OR2W3, OR1-1 |
Environmental signaling in fetus-mother interaction [12]
Environmental signaling in fetus-mother interaction [12] |
HeLa |
Human |
OR2A4 |
Regulation of actin cytoskeleton cytokinesis [44] |
Brain (and CNS in general) |
Rat, mouse
Mouse
Human
|
PSGR
MOR2.3, M71, C6, OR3
OLFR110/111, OLFR544
OR2L13, OR1E1, OR2J3, OR52L1, OR11H1
OR2H2, OR2A4, OR6K3 |
Not yet defined [33]
Possible involvement in developmental processes such as axon guidance [25], [45]
Brain physiology [46]
Cerebral cortex physiology [47]
Not yet defined [48] |
Lung
|
Human
|
OR51E2
OR1D2, OR2AG1 |
Modulation of airway smooth muscle (ASM) cells proliferation [49]
Regulation of pathophysiological processes [50] |
Table 1. Distribution of the main olfactory receptors identified in non-chemosensory tissues
In contrast to the OSN, however, ORs activation in those tissues is no longer associated to cell depolarization and neuronal impulse generation but linked to AC/cAMP or PLC/IP3 pathways. Ligand binding to the extracellular N-terminus domain induce a conformational change in the receptor which leads to a GDP/GTP binding exchange at Gα subunit level [51].In the AC/cAMP pathway, mainly regulating lipid metabolism, the Gα subunit dissociates from ßγ heterodimer and diffuses along the inner membrane surface towards the effector protein ACIII, activating it and allowing the production of cAMP, that in turns act on the Protein Kinase A (PKA), a tetrameric complex made up of two regulatory (R) and two catalytic (C) subunits. Binding of two molecules of cAMP to each of the R subunits, triggers PKA activation, which exerts its kinase activity on a large number of cytosolic and nuclear proteins [52]. Additionally, the cAMP can also activate CNG channels located on endoplasmic reticulum, increasing intracellular Ca++ concentrations. In the PLC/IP3 pathway, mostly involving, on the other hand, glucose metabolism, GTP-Gα subunit activates the effector protein Phospholipase C (PLC) which in turns hydrolyzes phosphatidylinositol 4,5-biphosphate (PIP2) – a constituent of the plasmatic membrane – in inositol 1,4,5-triphosphate (IP3) and 1,2-diacylglicerol (DAG), critical second messengers responsible for intracellular Ca++ signaling regulation [53] (Figure 2). Further on, because its intrinsic GTPase activity, the Gα subunit hydrolyses GTP in GDP and Pi, thereby inducing self-inactivation. Gα-GDP subunit associates again with the heterodimer ßγ and with the GPCR, bringing back the receptor to its inactive state.
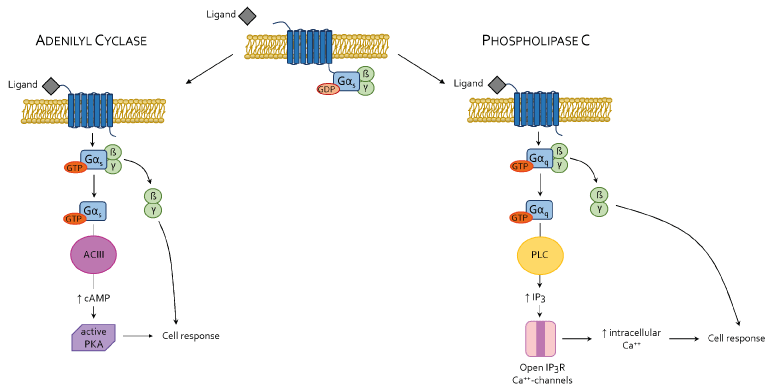
Figure 2. Transition from inactive (unbound ligand) to active (bound ligand) state of a typical GPCR. In ACIII/cAMP GPCR, ligand binding provokes conformational changes in the receptor, inducing a GDP/GTP exchange on Gαs subunit, its activation and dissociation from the αßγ heterotrimer and migration to ACIII. Active ACIII produces cAMP which activates PKA, resulting in phosphorylation and activation of different target proteins. In PLC/IP3 GPCR, ligand binding provokes conformational changes in the receptor, inducing a GDP/GTP exchange on Gαq subunit, its activation and dissociation from the αßγ heterotrimer and migration to PLC. Active PLC produces DAG and IP3 from PIP2 hydrolysis. IP3 in turns activates IP3 sensitive Ca++ channels present on endoplasmic reticulum (ER) membranes where Ca++ is stored, thus increasing intracellular Ca++ levels
Ectopically expressed ORs play a role in glucose and lipid metabolism: As mentioned above, many ORs have been detected outside of the MOE, although the function of the large majority of these receptors remains unclear. However, the presence of some ORs in in tissues and organs such as liver, kidneys, pancreas and adipose tissue, may suggest their involvement in the regulation glucose and lipid homeostasis. Up to now, it is possible to confirm the ability of some of those ectopically expressed ORs to exert regulatory function on blood glucose levels and lipid metabolism [54], opening the possibility for their exploitation in managing metabolic disorders such as type 2 diabetes mellitus (T2DM) and nonalcoholic fatty liver diseases (NAFLD) or nonalcoholic steatohepatitis (NASH) [55-57], i.e. a family of human disorders that develop in association with the Metabolic Syndrome [58] (Table 2).
OR |
Odorant(s)/ligand(s) |
Pathway(s) |
Main function(s) |
OLFR544 |
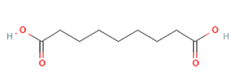 Azelaic acid Azelaic acid
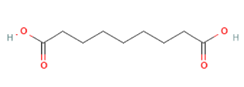 Azelaic acid Azelaic acid
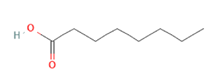 Octanoic acid Octanoic acid
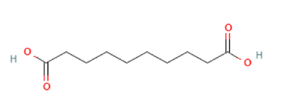 Sebacic acid Sebacic acid
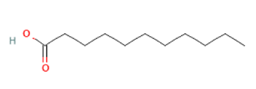 Undecanoic acid Undecanoic acid
|
cAMP/Ca++
cAMP/HSL
and
cAMP/CREB |
Ca++-dependent glucagon secretion [42]
HSL-induced triglycerides breakdown [38]
PPARα-induced FAO and inhibition of triglycerides synthesis via HES-1 repression [38] |
OR1A1/OLFR43 |
(-)-Carvone
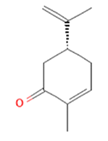
Geraniol
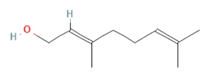
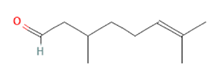 Citronellal Citronellal
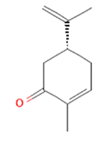 (-)-Carvone (-)-Carvone
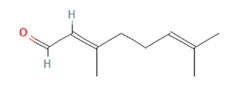 Citral Citral
 Nerol Nerol
3-Methyl-2,4-nonanedione
 |
cAMP/Ca++
cAMP/CREB |
GLP-1 and Ca++-dependent glucagon secretion [59], [60]
PPARα-induced FAO and inhibition of triglycerides synthesis via HES-1 repression [35], [61]–[63] |
OR51E1 |
 Nonanoic acid Nonanoic acid
|
cAMP/Ca++ |
Ca++-dependent GLP-1 secretion [43] |
OR10J5/OLFR16 |
 α-cedrene α-cedrene
|
cAMP-CREB
cAMP-HSL
cAMP-AMPK |
PPARα-induced FAO and inhibition of triglycerides synthesis via HES-1 repression, hepatic steatosis improvement [36], [64], [65]
HSL-induced triglycerides breakdown, hepatic steatosis improvement [36], [64]
LXRα and SREBP-1c repression, inhibition of lipogenic genes (ACC, FAS, SCD-1, hepatic steatosis improvement [36], [64], [65] |
OLFR15, OLFR821 |
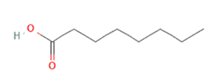 Octanoic acid Octanoic acid
|
PLC/IP3 |
Increased expression of GK, potentiation of glucose absorption and GSIS [27], [28]
|
OLFR734 |
Asprosin (endogenous) |
cAMP/CREB |
Increased expression of G6PC and PCK1, gluconeogenesis stimulation [34] |
OLFR1393 |
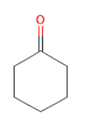 Cyclohexanone Cyclohexanone
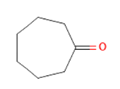 Cycloheptanone Cycloheptanone
Cycloheptanol
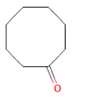 Cyclooctanone Cyclooctanone
|
cAMP-? |
Glucose reabsorption at renal tubules level via increasing expression of SGLT1 and SGLT2 [40] |
Table 2. State of the art of the main ORs involved in glucose and lipid metabolism, their relative odorant ligands and pathways activated.
ORs in glucose homeostasis
Glucose metabolism: Glucose is a 6-carbon sugar found in different foods both in simple (mono- and di-saccharides, such as fructose and lactose, respectively) and complex (e.g., starch) forms, and used as principal source of energy by our organism, reason why glucose transporters are present on surface membranes of all our cells [66]. Those transporters, classified in Sodium Glucose Transporters (SGLTs) and facilitated diffusion Glucose Transporters (GLUTs), are differently distributed among different cell types [67] and allow glucose uptake from blood circulation to (i) produce energy (ATP) via a series of biochemical reactions (glycolysis) or, in case of excess glucose, to (ii) store it under the form of glycogen (glycogen synthesis) in liver and skeletal muscles [68-70]. The balance between blood glucose, glucose absorption and glycogen deposition is finely regulated by the two peptide hormones insulin and glucagon, produced in Langerhans pancreatic islets, by ß- and α-cells respectively [71]. In a simple way: after a meal, blood glucose levels are increased (causing hyperglycemia) and insulin secretion is stimulated. After its release in the bloodstream, insulin reaches its main target tissues such as hepatic, skeletal and adipose tissues, binds to its receptor and increase translocation of GLUT4 transporter to the surface membrane, potentiating glucose uptake [72,73], as well as enhancing free fatty acids and amino acids uptake for lipogenesis and protein synthesis. These processes are furthermore potentiated by glucagon-like peptide-1 (GLP-1), released by gut enteroendocrine cells and responsible for increase of insulin secretion [74].
Conversely, during fasting periods glucagon [75], that is released by pancreatic cells, exerts several counter-regulatory effects including (i) reduction of hepatic glucose uptake, (ii) I breakdown of the previously stored glycogen (glycogenolysis) and, (iii) de novo glucose synthesis (gluconeogenesis) [76-78].
ORs and glucose metabolism in human disorders: In pathological conditions, glucose metabolism is altered generally due to an improper insulin production and subsequent long-lasting high blood glucose levels in T2DM, also known as adult-onset diabetes, is one of the most common widespread chronic metabolic disorders worldwide, accounting for almost the 90% of all diabetic patients [79]. T2DM is characterized by persistent hyperglycemia mainly due to defective insulin secretion by pancreatic ß-cells and/or loss of responsiveness of insulin-sensitive tissues, causing the development of insulin resistance (IR) and hyperinsulinemia onset [80]. Persistent high levels of blood glucose in non-treated patients are responsible for micro- and macro-vascular complications such as retinopathy, nephropathy and cardiovascular comorbidities [81-86].
Recent studies have confirmed that some ectopically expressed ORs exert a regulatory function in glucose metabolism. Olfr43 (homologous of the human OR1A1 gene) and OR51E1, expressed by L and K enteroendocrine cells and respectively activated by geraniol, citronellal and (-)-carvone or nonanoic acid binding, participates in glucose homeostasis by inducing GLP-1 secretion which, in turns, stimulates insulin production [43,59,60]. Thus, ORs add to the many GPCRs that regulate GLP-1 release from L cells. These promotes GLP1 secretion in response to various nutrients or microbial products and exert a critical role in regulating insulin secretion and sensitivity. L cells expressed various lipid sensing receptors, including the free fatty acid receptor (FFAR)2/GPR43 and FFAFA3/GPR41, two receptors for short chain fatty acids (SCFA) [87], along with FFAR1/GPR40 and FFAR40/GP120 two receptors for long chain fatty acids [88], GPR119 that is activated by fatty acid metabolites such as ethanololemanie such as N-acylethanolamines and 2-monoacylglycerols [89], and GPBAR1 (also known as TGR5) a receptor for secondary bile acids litocholic acid (LCA) and deoxycholic acid (DCA), generated from the intetsinal microbiota [90]. These array of receptors are mostly known for their ability to sense microbial product, and, in addition to ORs provide an intricated network of chemical signaling that integrate nutrients, intstinal microbiota and host metabolism and immune system, that could be exploited for therapeutic purposes [91-93].
To further highligt these interactions, Olfr15 and Olfr821, detected in ß-pancreatic islets and ß-cell line MIN6, are capable to recognize and bind the medium-chain saturated fatty acid (MCFA) octanoic acid (also known as caprylic acid, naturally found in coconut and palm oils [94]), thus activating downstream PLC/IP3 pathway, inducing glucokinase (GK) expression and potentiating glucose absorption and glucose-stimulated insuline secretion (GSIS) [27,28]. In both cases, activation of those receptors exerts an hypoglycemic and anti-diabetic function, finally lowering blood glucose levels via increasing insulin secretion.
On the other hand, different ORs have been linked to be directly involved in diabetes pathogenesis. In particular, Olfr1393, expressed in the kidneys by the proximal tubules, has been confirmed to be involved in glucose reabsorption at renal level by increasing expression and membrane translocation of the sodium-glucose cotransporters SGLT1 and SGLT2 [40], thus increasing blood glucose levels and emerging indeed as an important contributor to T2DM development. Despite the molecular mechanism is not so clear, further studies have confirmed Olfr1393 capability to modulate SGLT1 and SGLT2: infact, SGLT2 expression was reduced in Olfr1393 KO mice, resulting in improvement of hyperglycemia and glucose tolerance [95,96]. Both in vitro and in vivo studies revealed and increased expression of SGLT2, and thus of glucose reabsorption, in T2DM patients [97]: although not yet completely understood, it seems that SGLT2 overexpression should be due to a hyperphosphorilaton guided by insulin via PKA and PKC [98,99]. Conversely, SGLT2 genetic mutations and/ore deletions have been associated to familial renal glicosuria (FRG), with persistent high levels of glucose in the urine [100]. Importantly, SGLT2 inhibitors (i.e., canagliflozin, dapagliflozin, and empagliflozin) have been shown effective in the treatment of T2DM by reducing glucose reabsorption in the proximal renal tubule thus lowering blood glucose levels and glucose toxicity [101-103]. SGLT2 inhibitors have gain approval for the treatment of T2DM patients [104,105] and targeting Olfr1393 thus preventing SGLT2 expression offers novel therapeutic possibilities in tyhis setting. Olfr734, that is expressed by hepatocytes, is activated by asprosin, an endogenous fasting-induced hormone produced by adipose tissues [106], that in its turn incraeses the expression of glucose-6-phosphatase (G6PC) and phospho-enolpyruvate carboxykinase (PCK1) thus promoting gluconeogenesis and blood glucose release as well as stimulating appetite at hypotalamic level [34]. The expression of Olfr543, Olfr544, Olfr545 and Olfr1349 has been detected also in the pancreatic α-cells, but only the stimulation of Olfr544 by azelaic acid results in Ca++-dependant glucagon secretion [42] (Figure 3).
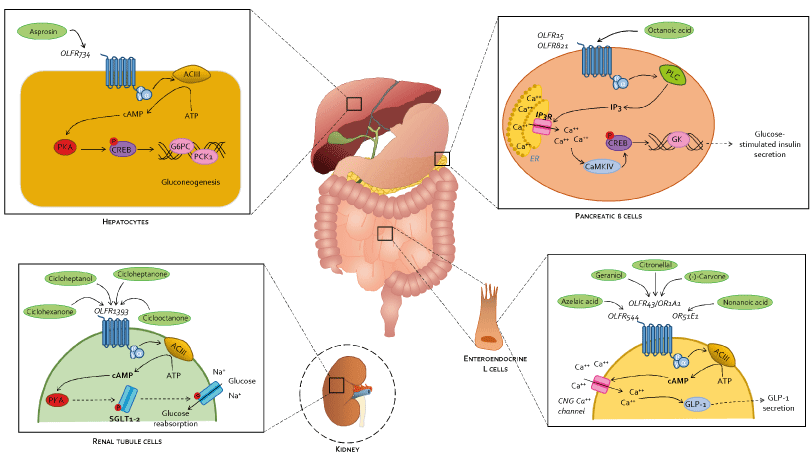
Figure 3. Modulation of glucose metabolism by ectopically expressed ORs via cAMP/CREB and PLC/IP3 pathways. In the liver, PKA-activated CREB induces the expression of G6PC and PCK1 genes, indeed promoting gluconeogenesis. In renal tubule cells, PKA seems to be involved in SGLT1-2 phosphorylation and subsequent membrane translocation, potentiating renal glucose reabsorption. In enteroendocrine L cells, cAMP acts directly on activating CNG Ca++ channels: increase intracellular Ca++ induces GLP-1 secretion. In pancreatic β-cells, IP3 binds IP3 receptors (IP3R) located on endoplasmic reticulum (ER) where intracellular Ca++ is stored, inducing their opening and cytoplasmic Ca++ increase. Ca++ activates calcium/calmodulin-dependent protein kinase IV (CaMKIV), subsequent CREB phosphorylation and activation and GK gene expression, resulting in final glucose-stimulated insulin secretion
ORs and lipid metabolism
Lipid homeostasis: Lipids are an eterogeneous group of hydrophobic molecules majorly belonging to fatty acids (FAs), triglycerides (TGs), phospholipids, steroids and glycolipids classes [107] all necessary for our organism due to their implications in many different aspects of cellular homeostasis and metabolism. Cholesterol, is the starting molecule in the biosynthesys of hormones such as progesterone, aldosterone, cortisol and testosterone, other than be a fundamental costituent of cell membranes[108,109] together with phospholipids [110], regulating important aspects of the bilayer such as fluidity, permeability and rigidity [111]. Glycolipids are important membrane constituent as well due to their role in facilitating cell-cell interactions [112, 113]. However, almost the 90% of the lipid intake in a normal diet is represented by triglycerides, suppling most of the lipidic energy source [114]. After enzymatic digestion (borne by lingual, gastric and pancreatic lipases) and instestinal bile salts emulsion, FFAs, free cholesterol and 2-monoacylglycerols are the main products ready to be absorbed by enterocytes under the form of micelles [115]. Once inside the cell, FFAs and 2-monoacylglycerols are converted again in TGs, cholesterol is esterified and together are incorporated in small particles named chylomicrons. Those chylomicrons, constituted up to 90% by TGs, are reversed into the lymphatic system and transported to (i) skeletal and cardiac tissues, where TGs are released to be used for energy production or to (ii) the adipose tissue to be stored as tryglycerides [116]. After TGs release, what remains of chylomicrons is absorbed by hepatocytes to form very low-density lipoproteins (VLDLs, costituted up to 60% by TGs), to bring lipids from the liver to peripeheral tissues. TGs depletion converts VLDLs in low-density lipoproteins (LDLs), predominantly made up of cholesterol which can be brought to peripheral tissues or, thanks to the activity of high-density lipoproteins (HDLs), back to the liver for biliary acids synthesis or to steroidogenics cells for hormones production [117]. Role of HDLs in fundamental in cholesterol homeostasis since by removing excess cholesterol molecules from the periphery (including atherosclerotic plaques) can be associated to a reduction of cardiovascular risk [118, 119].
During fasting periods, in glucose shortage, FFAs represents the primary source of energy for [120]: via the mitochondrial ß-oxidation pathway, fatty acids are converted in Acetyl CoA, which enters tricarboxylic acid cycle (TCA, also know as Krebs cycle) to finally obtain NADH and FADH2 molecules, used as final electron acceptor for ATP production [121]. At the same time, reduction of the ATP/ADP ratio induce phosphorilation and activation of AMPK [122], inhibiting lipogenesis by preventing expression of lipogenic genes [123], reducing hepatic lipid accumulation [124] and potentiating fatty acids oxidation via PPARα transcription factor [125].
Cholesterol and FAs are are not just introduced with the diet, but are also synthesized de novo in the liver starting from Acetyl CoA: sterol regulatory element binding protein (SREBP)-1a, -1c and -2 is activated in presence of low sterols levels, inducing the transciptional activation and expression of genes involved in FAs and cholesterol biosynthesis like fatty acid synthase (FAS), stearoyl-CoA desaturase (SCD1), mitochondrial glycerol-3-phosphate acyltransferase (mtGPAT), acetyl-CoA carboxylase (ACC) [126], and simultaneously decreasing ß-oxidation. Alongside, high levels of glucose activate PPARγ resulting in de novo lipogenesis and hepatic lipid deposition [127].
ORs and NAFLD/NASH
In pathological conditions, the balance between de novo lipogenesis and fatty acids ß-oxidation can be altered, resulting in dyslipidemia, excessive lipid accumulation and an increase risk for atherosclerosis and cardiovascular diseases [128]. Nonalcoholic fatty liver disease (NAFLD, also known as fatty liver or hepatic steatosis) is characterized by the abnormal deposition of lipids within the hepatic tissue and represents nowadays one of the most diffuse liver diseases in developed countries, affecting about the 30% of the US population. Anomalous storage of triglycerides in hepatocytes as a result of the imbalance in lipid metabolism (increased de novo lipogenesis, adipose tissue lipolysis and free fatty acids uptake vs impaired lipid ß-oxidation), owing to hepatic lipotoxicity, can induce NAFLD progression and worsening in nonalcoholic steatohepatitis (NASH), marked by the onset of inflammation and fibrosis, till cirrhosis and, at worst, hepatocellular carcinoma [129,130]. Typical NAFLD patient’s features are represented by IR, increased plasma triglycerides, high levels of circulating AST and ALT, abdominal obesity and fatty liver [131,132]. As seen also for T2DM, physical activity and healthy diet (with special reference to Mediterranean and ketogenic diets) are the starting point for ameliorating lipid homeostasis in NAFLD patient [133]. In addition to this, although no approved treatments are currently available for NAFLD/NASH, animal data and recent human trials seem to identify statins (e.g., atorvastatin, fluvastatin and rosuvastatin) as possible drugs to be used in NAFLD/NASH patients, giving beneficial results in terms of hyperlipidemia improvement, liver steatosis and inflammatory decrease, anti-fibrotic effect, as well as reduction for cardiovascular diseases risk and hepatocarcinoma [134-140]. Also, agonists of PPARα nuclear receptors could be used, thanks to their ability to induce mitochondrial ß-oxidation and reduce lipogenesis, finally resulting in decreased liver steatosis [127,141,142]. Liver-x-receptor α (LXRα), particularly expressed in metabolic tissues such as liver and adipocytes, is a nuclear transcriptional receptor mainly activated by cholesterol-derived endogenous ligands (e.g., oxysterols) which induces SREBP-1c and all its derived lipogenic genes expression [143]: for this reason, treatment with LXRα antagonist might be beneficial for NAFLD treatment by suppressing the activity of its target genes and alleviating lipogenesis [144-146].
During last years, several ORs have been investigated for their potential role in regulating lipid homeostasis in hepatic steatosis and lipolysis and, as a consequence, could be considered potential targets in the development of new drugs for NAFLD prevention [147]. ORs activation in hepatic and adipose tissue is linked to cAMP production and PKA activation resulting, finally, in phosphorylation and activation of three principal target proteins massively involved in lipid metabolism: hormone-sensitive lipase (HSL), cAMP response element binding protein (CREBP) and AMP-activated protein kinase (AMPK) [148] (Figure 3). Each of these different pathways (i.e., cAMP/HSL, cAMP/CREB and cAMP/AMPK) can be activated by binding of various ligands to specific ORs.
Stimulation and activation of mouse OLFR16 and its human homolog OR10J5, both expressed in liver, adipose and muscular tissues, using the sesquiterpene α-cedrene (a natural odorant compound found in cedarwood essential oils [149]), resulted in modulation of lipid metabolism via all the three different pathways previously cited [65,149,150]. Indeed, ligand-receptor binding induces phosphorylation and activation of HSL, by increasing triglycerides hydrolyzation, as well as CREB activation, with subsequent decreased expression of HES-1 and PPAR-γ resulting in reduction of triglycerides, cholesterol and free fatty acids in hepatocytes [151,152]. In parallel, CREB phosphorylation induce the expression of peroxisome proliferator-activated receptor γ coactivator1-α (PGC-1α) and its downstream thermogenic genes such as UCP1, involved in thermogenesis, energy balance and control of body weight [65,153-155]. Moreover, PKA-induced p-AMPK inhibits in-vitro LXRα expression, leading to SREBP-1c and its target genes downregulation, such as ACC, FAS and SCD-1, and alleviates in-vivo hepatic steatosis in mice fed HFD [36,64]. It has already been confirmed that inactivation of both SREBP-1c and SREPB-1a phosphorylation prevents the onset of fatty liver disease in mice, avoiding FAs and cholesterol synthesis [156,157]. Moreover, even if characterized by a lower binding affinity with respect to the natural α-cedrene, the synthetic lyral derivatives also binds and activates OLFR16 and OR10J5 [158]. In addition, current studies are still ongoing to unveil the specific endogenous ligand compound that seems to be released by injured muscle tissues and involved in cell adhesion and migration [32].
Targeting OLFR544 with azelaic acid (AzA), a non-toxic dicarboxylic acid naturally found in wheat, rye and barley [159], results in similar biological effects by acting via the cAMP/HSL and cAMP/CREB pathways. Highly expressed both in liver and adipose tissue, the activated receptor participates in regulation of cell metabolism, inducing transcriptional repression of genes involved in lipogenesis. AzA was confirmed to be able to reduce and revert adiposity [39] in mice fed HFD as a consequence of (i) increased triglycerides hydrolyzation (cAMP/HSL) and (ii) PPAR-α-induced fatty acid oxidation (FAO) and ketogenesis (cAMP/CREB) [38], together with AzA ability to restore hepatic markers to values close to normal [160,161]. Despite azelaic acid results to be the strongest agonist capable of activating OLFR544, octanoic acid, sebacic acid and undecanoic acid can activate the receptor, too, even though in a milder way [162].
cAMP/CREB pathway is also triggered when (-)-Carvone, a monoterpene presents in high quantity in fennel, caraway and spearmint essential oils [163], binds and activates the mouse OLFR43 and its human homolog OR1A1, both strongly expressed in the liver [35]. In-vitro decrease of intracellular triglycerides levels and lipid accumulation was furthermore confirmed by in-vivo reduction of hepatic steatosis [61], together with reduced expression of lipogenic genes such as PPAR-γ and SCD1 [62]. Besides recognizing (-)-carvone, it has been established OR43/OR1A1 binding ability towards about 30 different compounds among esters, terpenes (e.g., citral), alcohols (e.g., nerol), aldehydes and ketones (e.g., 3-Methyl-2,4-nonanedione) [63,164] (Figure 4).
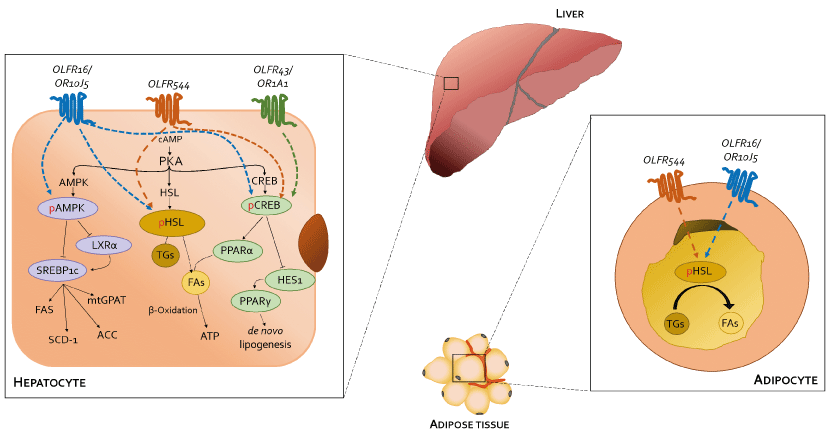
Figure 4. Modulation of lipid metabolism in hepatocytes and/or adipocytes by activation of ectopically expressed ORs through cAMP/HSL, cAMP/CREB and cAMP/AMPK pathways. In the former case, PKA phosphorylates and activates HSL, enzyme responsible for triglycerides decomposition, thus enhancing fatty acids β-oxidation. In the second case, PKA-dependent CREB phosphorylation indirectly inhibits PPARγ via HES1 inhibition thus reducing triglycerides synthesis on the one hand and inducing PPARα expression and, thus, stimulating fatty acids β-oxidation, on the other hand. In the latter case, active p-AMPK down-regulates LXRα and RXR, thus inhibiting expression of the key lipogenic transcription factor SREBP-1c and all its downstream genes, such as aP2, FAS, SCD1, ACC, and mtGPAT, finally reducing lipogenesis
Interestingly, three new olfactory receptors (i.e., OR4C11, OR4P4 and OR4S2) have been detected in the adipose tissue and, as demonstrated in different linkage analysis, their disruption (associated to the deletion of the 11q11 region where they are mapped) leads to fat accumulation, confirming them as new risk factors for obesity [165] and corroborating the hypothesis that ORs might have a protective role against fat accumulation in hepatic and adipose tissue [166]. Another genome-wide associated study (GWAS) allowed to confirm the 11q11 region as a copy number variation region (CNVR) associated to early-onset extreme obesity, confirming reduced number of CNV of that region [167]. Anyway, further studies are needed to unveil the molecular mechanism which underlies beyond the activation of these antiobesogenic ORs.
Conclusions
T2DM and NAFLD commonly exist together in the so-called Metabolic Syndrome (MetS) [168,169]. Atherogenic dyslipidemia, abdominal obesity, hypertension and insulin resistance are typical clinical features of the MetS and represent all together a cluster of risk factors (RFs) for the development of cardiovascular diseases (CVDs) [170]. Therefore, managing to control and restore glucose and lipid metabolism back to normality is fundamental to treat symptomatology and pathophysiology of T2DM and NAFLD and solve the burden of the Metabolic Syndrome.
Already well-known in literature is the onset of olfactory impairment in diabetic and obese patients [171], unveiling the connection between olfaction (i.e., ORs), glucose and lipid metabolism. Not only: day after day, an increasing number of ectopically expressed ORs are discovered, together with their ability to take part in glucose and lipid metabolism. Thus, a deepened knowledge is required in order to understand how those ORs work on modulating glucose and lipid metabolism, what pathways they are able to trigger and what are the biological effects induced by their activation.
As previously mentioned, diverse ORs can activate different intracellular signaling pathways, leading to different biological effects; moreover, triggering the same receptor with the same ligand makes possible to activate simultaneously multiple pathways and even if the function of the large majority of them is still unclear, it has been possible to link some of them to specific pathways regulating glucose and lipid metabolism. Activation of Olfr43/OR1A1, OR51E1, Olfr15 and Olfr821 such as inhibition of Olfr734 and Olfr1393 can induce kidney hyperfiltration, renal glucose reabsorption and blood glucose levels reduction, giving relieve to diabetic patients. At the same time, triggering Olfr16/OR10J5 and Olfr43/OR1A1 while inhibiting OR4C11, OR4P4 and OR4S2 potentiate lipids breakdown and adiposity reduction, being a possible successful approach for patients affected by NAFLD. Nevertheless, Olfr544 results to be an ambiguous target: indeed, its activation could be useful to induce triglycerides hydrolyzation but unfavorable due to the subsequent blood glucose increase.
Therefore, it is possible to infer that (i) the large majority of odorants known so far are ORs agonists both in glucose and lipid metabolism regulation and (ii) they mainly belong to two types of biological molecules, i.e., saturated fatty acids and terpenes/terpenoids, indicating those two compound classes of primary interest in the continuous search for new natural ligands.
Regrettably, only for some of the above mentioned ORs it has been possible to define an endogenous ligand (e.g., Asprosin for OLFR734) or exogenous one; indeed, the great major of them (about the 80%) are identified as “orphan receptors”, that is why further and more in-depth studies are still needed to define new ORs odorants and their biological role in cell metabolism.
Moreover, the large majority of those studies have been developed by using mouse models that, despite their genetic and physiological similarities with the human species and their diffuse use for human diseases modeling [172], still may have some differences in term of tissue-specific ORs expression or different ability to trigger intracellular signaling pathways.
Finally, taking in consideration all these aspects, it is clearly affirmable an ORs’ involvement in glucose and lipid metabolism. Despite that, additional studies are still needed to find out new ORs and respective odorants, as well as their possible targeting for the treatment of diseases marked by dysregulation in glucose and lipid metabolism such as T2DM and NAFLD, with the final aim to relieve hypertension, dyslipidemia, obesity, hyperglycemia and insulin resistance, typical clinical features of the Metabolic Syndrome.
References
- Buck L and Axel R (1991) A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65: 175-187. [Crossref]
- Sharma A, Kumar R, Aier I, Semwal R, Tyagi P, et al. (2019) Sense of Smell: Structural, Functional, Mechanistic Advancements and Challenges in Human Olfactory Research. Curr Neuropharmacol 17: 891-911. [Crossref]
- Buck LB (1992) The olfactory multigene family. Curr Opin Genet Dev 2: 467-473.
- De Munnik SM, Smit MJ, Leurs R, Vischer HF (2015) Modulation of cellular signalling by herpesvirus-encoded G protein-coupled receptors. Front Pharmacol 6: 40. [Crossref]
- Beigi M and Zell A (2006) A Novel Method for Classifying Subfamilies and Sub-subfamilies of G-Protein Coupled Receptors BT-Biological and Medical Data Analysis. 25-36.
- Bowman GL (2017) Biomarkers for early detection of Parkinson disease: A scent of consistency with olfactory dysfunction. Neurology 89: 1432-1434.
- Kang N and Koo J (2012) Olfactory receptors in non-chemosensory tissues. BMB Rep 45: 622.
- Malnic B, Godfrey PA, Buck LB (2004) The human olfactory receptor gene family. Proc Natl Acad Sci U. S. A. 101: 2584 LP- 2589.
- Maßberg D and Hatt H (2018) Human Olfactory Receptors: Novel Cellular Functions Outside of the Nose. Physiol Rev 98: 1739-1763.
- Parmentier M (1992) Expression of members of the putative olfactory receptor gene family in mammalian germ cells. Nature 355: 453-455. [Crossref]
- Drutel G, Arrang JM, Diaz J, Wisnewsky C, Schwartz K, et al. (1995) Cloning of OL1, a putative olfactory receptor and its expression in the developing rat heart. Receptors Channels 3: 33-40.
- Itakura S, Ohno K, Ueki T, Sato K, Kanayama N, et al. (2006) Expression of Golf in the rat placenta: Possible implication in olfactory receptor transduction. Placenta 27: 103-108. [Crossref]
- Pluznick JL (2009) Functional expression of the olfactory signaling system in the kidney. Proc Natl Acad Sci. U. S. A 106: 2059-2064.
- Pluznick JL (2013) Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc. Natl. Acad. Sci. U. S. A 110:. 4410-4415.
- Braun T, Voland P, Kunz L, Prinz C, Gratzl M, et al. (2007) Enterochromaffin cells of the human gut: sensors for spices and odorants. Gastroenterology 132: 1890-1901.
- Kurtz R, Steinberg LG, Betcher M, Fowler D, Shepard BD (2020) The Sensing Liver: Localization and Ligands for Hepatic Murine Olfactory and Taste Receptors. Front. Physiol 11: 574082. [Crossref]
- Tong T, Wang Y, Kang SG, Huang K (2021) Ectopic Odorant Receptor Responding to Flavor Compounds: Versatile Roles in Health and Disease. Pharm 13(8).
- Spehr M, Gisselmann G, Poplawski A, et al. (2003) Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science 299(5615):2054-2058.
- Flegel C, Vogel F, Hofreuter A, et al. (2016) Characterization of the Olfactory Receptors Expressed in Human Spermatozoa. Front Mol Biosci 2.
- Fukuda N, Touhara K (2006) Developmental expression patterns of testicular olfactory receptor genes during mouse spermatogenesis. Genes Cells 11(1):71-81. [Crossref]
- Fukuda N, Yomogida K, Okabe M, Touhara K (2004) Functional characterization of a mouse testicular olfactory receptor and its role in chemosensing and in regulation of sperm motility. J Cell Sci 117(Pt 24):5835-5845. [Crossref]
- Gaudin JC, Breuils L, Haertlé T (2001) New GPCRs from a Human Lingual cDNA Library. Chem Senses 26(9):1157-1166.
- Durzyński L, Gaudin JC, Myga M, Szydłowski J, et al. (2005) Olfactory-like receptor cDNAs are present in human lingual cDNA libraries. Biochem Biophys Res Commun 333: 264-272. [Crossref]
- Jovancevic N, Dendorfer A, Matzkies M, et al. (2017) Medium-chain fatty acids modulate myocardial function via a cardiac odorant receptor. Basic Res Cardiol 112:13.
- Weber M, Pehl U, Breer H, Strotmann J (2002) Olfactory receptor expressed in ganglia of the autonomic nervous system. J Neurosci Res 68:176-184.
- Blache P, Gros L, Salazar G, Bataille D (1998) Cloning and tissue distribution of a new rat olfactory receptor-like (OL2). Biochem Biophys Res Commun 242: 669-672. [Crossref]
- Leem J, Shim HM, Cho H, Park JH (2018) Octanoic acid potentiates glucose-stimulated insulin secretion and expression of glucokinase through the olfactory receptor in pancreatic β-cells. Biochem Biophys Res Commun 503(1):278-284.
- Munakata Y, Yamada T, Imai J, et al. (2018) Olfactory receptors are expressed in pancreatic β-cells and promote glucose-stimulated insulin secretion. Sci Rep 8: 1499-19765
- Feingold EA, Penny LA, Nienhuis AW, Forget BG. (1999) An olfactory receptor gene is located in the extended human beta-globin gene cluster and is expressed in erythroid cells. Genomics 61: 15-23.
- Xu LL, Stackhouse BG, Florence K, et al. (2000) PSGR, a novel prostate-specific gene with homology to a G protein-coupled receptor, is overexpressed in prostate cancer. Cancer Res 60: 6568-6572. [Crossref]
- Neuhaus EM, Zhang W, Gelis L, Deng Y, Noldus J, et al. (2009) Activation of an olfactory receptor inhibits proliferation of prostate cancer cells. J Biol Chem 284: 16218-16225. [Crossref]
- Griffin CA, Kafadar KA, Pavlath GK. (2009) MOR23 promotes muscle regeneration and regulates cell adhesion and migration. Dev Cell 17: 649-661.
- Yuan TT, Toy P, McClary JA, Lin RJ, Miyamoto NG, Kretschmer PJ. (2001) Cloning and genetic characterization of an evolutionarily conserved human olfactory receptor that is differentially expressed across species. Gene 278: 41-51.
- Li E, Shan H, Chen L, et al. (2019) OLFR734 Mediates Glucose Metabolism as a Receptor of Asprosin. Cell Metab 30: 319-328.e8.
- Wu C, Jia Y, Lee JH, et al. (2015) Activation of OR1A1 suppresses PPAR-γ expression by inducing HES-1 in cultured hepatocytes. Int J Biochem Cell Biol 64:75-80. [Crossref]
- Tong T, Ryu SE, Min Y, et al. (2017) Olfactory receptor 10J5 responding to α-cedrene regulates hepatic steatosis via the cAMP–PKA pathway. Sci Rep 7: 9471.
- Maßberg D, Simon A, Häussinger D, et al. (2015) Monoterpene (-)-citronellal affects hepatocarcinoma cell signaling via an olfactory receptor. Arch Biochem Biophys 566: 100-109.
- Wu C, Hwang SH, Jia Y, et al. (2017) Olfactory receptor 544 reduces adiposity by steering fuel preference toward fats. J Clin Invest 127: 4118-4123. [Crossref]
- Crunkhorn S. (2017) Ectopic olfactory receptor activation reverses obesity. Nat Rev Drug Discov 16: 826-827.
- Shepard BD, Cheval L, Peterlin Z, et al. (2016) A Renal Olfactory Receptor Aids in Kidney Glucose Handling. Sci Rep 6: 35215. [Crossref]
- Rajkumar P, Aisenberg WH, Acres OW, Protzko RJ, Pluznick JL, et al. (2014) Identification and characterization of novel renal sensory receptors. PLoS One 9: e111053.
- Kang N, Bahk YY, Lee N, et al. (2015) Olfactory receptor Olfr544 responding to azelaic acid regulates glucagon secretion in α-cells of mouse pancreatic islets. Biochem Biophys Res Commun 460: 616-621.
- Han YE, Kang CW, Oh JH, et al. (2018) Olfactory Receptor OR51E1 Mediates GLP-1 Secretion in Human and Rodent Enteroendocrine L Cells. J Endocr Soc 2: 1251-1258. [Crossref]
- Zhang X, Bedigian A V, Wang W, Eggert US. (2012) G protein-coupled receptors participate in cytokinesis. Cytoskeleton (Hoboken) 69: 810-818.
- Otaki JM, Yamamoto H, Firestein S. (2004) Odorant receptor expression in the mouse cerebral cortex. J Neurobiol 58: 315-327.
- Gaudel F, Stephan D, Landel V, Sicard G, Féron F, et al. (2019) Expression of the Cerebral Olfactory Receptors Olfr110/111 and Olfr544 Is Altered During Aging and in Alzheimer’s Disease-Like Mice. Mol Neurobiol 56: 2057-2072.
- Garcia-Esparcia P, Schlüter A, Carmona M, et al. (2013) Functional Genomics Reveals Dysregulation of Cortical Olfactory Receptors in Parkinson Disease: Novel Putative Chemoreceptors in the Human Brain. J Neuropathol Exp Neurol 72: 524-539.
- Ferrer I, Garcia-Esparcia P, Carmona M, et al. (2016) Olfactory Receptors in Non-Chemosensory Organs: The Nervous System in Health and Disease. Front Aging Neurosci 8: 163. [Crossref]
- Aisenberg WH, Huang J, Zhu W, et al. (2016) Defining an olfactory receptor function in airway smooth muscle cells. Sci Rep 6: 38231.
- Kalbe B, Knobloch J, Schulz VM, et al. (2016) Olfactory Receptors Modulate Physiological Processes in Human Airway Smooth Muscle Cells. Front Physiol 7: 339. [Crossref]
- Marinissen MJ, Gutkind JS. (2001) G-protein-coupled receptors and signaling networks: emerging paradigms. Trends Pharmacol Sci 22: 368-376.
- Sassone-Corsi P (2012) The cyclic AMP pathway. Cold Spring Harb Perspect Biol 4(12).
- Sekiya F (2013) Phospholipase C. In: Lennarz WJ, Lane MDBTE of BC (Second E, eds. Academic Press; 467-471).
- Ravnskjaer K, Madiraju A, Montminy M (2016) Role of the cAMP Pathway in Glucose and Lipid Metabolism. Handb Exp Pharmacol 233: 29-49. [Crossref]
- Eckel RH, Grundy SM, Zimmet PZ (2005) The metabolic syndrome. Lancet 365: 1415-1428.
- Saklayen MG. (2018) The Global Epidemic of the Metabolic Syndrome. Curr Hypertens Rep 20: 12.
- Samson SL, Garber AJ. (2014) Metabolic syndrome. Endocrinol Metab Clin North Am 43: 1-23.
- Lonardo A, Ballestri S, Marchesini G, Angulo P, Loria P. (2015) Nonalcoholic fatty liver disease: a precursor of the metabolic syndrome. Dig liver Dis Off J Ital Soc Gastroenterol Ital Assoc Study Liver 47: 181-190. [Crossref]
- Kim KS, Lee IS, Kim KH, et al. (2017) Activation of intestinal olfactory receptor stimulates glucagon-like peptide-1 secretion in enteroendocrine cells and attenuates hyperglycemia in type 2 diabetic mice. Sci Rep 7: 13978.
- Jones B, Bloom SR, Buenaventura T, Tomas A, Rutter GA (2018) Control of insulin secretion by GLP-1. Peptides 100: 75-84.
- Wu C, Thach TT, Kim YJ, Lee SJ (2019) Olfactory receptor 43 reduces hepatic lipid accumulation and adiposity in mice. Biochim Biophys acta Mol cell Biol lipids 1864: 489-499. [Crossref]
- Alsanea S, Liu D (2017) BITC and S-Carvone Restrain High-Fat Diet-Induced Obesity and Ameliorate Hepatic Steatosis and Insulin Resistance. Pharm Res 34: 2241-2249.
- Geithe C, Noe F, Kreissl J, Krautwurst D (2017) The Broadly Tuned Odorant Receptor OR1A1 is Highly Selective for 3-Methyl-2,4-nonanedione, a Key Food Odorant in Aged Wines, Tea, and Other Foods. Chem Senses 42: 181-193.
- Tong T, Yu R, Park T (2019) α-Cedrene protects rodents from high-fat diet-induced adiposity via adenylyl cyclase 3. Int J Obes (Lond) 43: 202-216. [Crossref]
- Tong T, Park J, Moon C, Park T. (2018) Regulation of Adipogenesis and Thermogenesis through Mouse Olfactory Receptor 23 Stimulated by α-Cedrene in 3T3-L1 Cells. Nutrients 10(11).
- Gurung P, Jialal I (2022) Plasma Glucose
- Navale AM, Paranjape AN (2016) Glucose transporters: physiological and pathological roles. Biophys Rev 8: 5-9.
- Desai MS, Shabier Z, Taylor M, et al. (2010) Hypertrophic cardiomyopathy and dysregulation of cardiac energetics in a mouse model of biliary fibrosis. Hepatology 51: 2097-2107. [Crossref]
- Adeva-Andany MM, González-Lucán M, Donapetry-García C, Fernández-Fernández C, Ameneiros-Rodríguez E, et al. (2016) Glycogen metabolism in humans. BBA Clin 5: 85-100.
- Panda S. (2016) Circadian physiology of metabolism. Science 354: 1008-1015.
- Han HS, Kang G, Kim JS, Choi BH, Koo SH, et al. (2016) Regulation of glucose metabolism from a liver-centric perspective. Exp Mol Med 48: e218. [Crossref]
- Furtado LM, Somwar R, Sweeney G, Niu W, Klip A, et al. (2002) Activation of the glucose transporter GLUT4 by insulin. Biochem Cell Biol 80: 569-578.
- Kahn CR. (1985) The molecular mechanism of insulin action. Annu Rev Med 36: 429-451.
- Drucker DJ. (2018) Mechanisms of Action and Therapeutic Application of Glucagon-like Peptide-1. Cell Metab 27: 740-756.
- Ramnanan CJ, Edgerton DS, Kraft G, Cherrington AD (2011) Physiologic action of glucagon on liver glucose metabolism. Diabetes Obes Metab 13 Suppl 1(Suppl 1): 118-125. [Crossref]
- Nordlie RC, Foster JD, Lange AJ (1999) Regulation of glucose production by the liver. Annu Rev Nutr 19: 379-406.
- Janah L, Kjeldsen S, Galsgaard KD, et al. (2019) Glucagon Receptor Signaling and Glucagon Resistance. Int J Mol Sci 20(13).
- Jiang G, Zhang BB (2003) Glucagon and regulation of glucose metabolism. Am J Physiol Endocrinol Metab 284: E671-E678. [Crossref]
- Chatterjee S, Khunti K, Davies MJ. (2017) Type 2 diabetes. Lancet (London, England) 389: 2239-2251.
- Lomberk G, Urrutia R. (2009) Primers on Molecular Pathways —The Insulin Pathway. Pancreatology 9: 203-205.
- Galicia-Garcia U, Benito-Vicente A, Jebari S, et al. (2020) Pathophysiology of Type 2 Diabetes Mellitus. Int J Mol Sci 21(17).
- DeFronzo RA, Ferrannini E, Groop L, et al. (2015) Type 2 diabetes mellitus. Nat Rev Dis Prim 1: 15019.
- Niswender KD (2011) Basal insulin: physiology, pharmacology, and clinical implications. Postgrad Med 123: 17-26. [Crossref]
- Pivari F, Mingione A, Brasacchio C, Soldati L (2019) Curcumin and Type 2 Diabetes Mellitus: Prevention and Treatment. Nutrients 11(8).
- Corcoran C, Jacobs TF (2022) Metformin.
- Standards of Medical Care in Diabetes (2019) Abridged for Primary Care Providers. Clin Diabetes 37: 11-34.
- Tang C, Ahmed K, Gille A, et al. (2015) Loss of FFA2 and FFA3 increases insulin secretion and improves glucose tolerance in type 2 diabetes. Nat Med 21: 173-177. [Crossref]
- Grundmann M, Bender E, Schamberger J, Eitner F (2021) Pharmacology of Free Fatty Acid Receptors and Their Allosteric Modulators. Int J Mol Sci 22(4).
- Chimerel C, Emery E, Summers DK, Keyser U, Gribble FM, et al. (2014) Bacterial metabolite indole modulates incretin secretion from intestinal enteroendocrine L cells. Cell Rep 9: 1202-1208.
- iorucci S, Distrutti E, Carino A, Zampella A, Biagioli M (2021) Bile acids and their receptors in metabolic disorders. Prog Lipid Res 82: 101094.
- Fiorucci S, Distrutti E. (2021) Linking liver metabolic and vascular disease via bile acid signaling. Trends Mol Med. Published online November.
- Kim MH, Kang SG, Park JH, Yanagisawa M, Kim CH, et al. (2013) Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 145: 310-396.
- Fiorucci S, Carino A, Baldoni M, et al. (2021) Bile Acid Signaling in Inflammatory Bowel Diseases. Dig Dis Sci 66: 674-693. [Crossref]
- Lemarié F, Beauchamp E, Legrand P, Rioux V (2016) Revisiting the metabolism and physiological functions of caprylic acid (C8:0) with special focus on ghrelin octanoylation. Biochimie 120: 40-48.
- Shepard BD, Koepsell H, Pluznick JL (2019) Renal olfactory receptor 1393 contributes to the progression of type 2 diabetes in a diet-induced obesity model. Am J Physiol Renal Physiol 316: F372-F381.
- Schiazza AR, Considine EG, Betcher M, Shepard BD (2021) Loss of renal olfactory receptor 1393 leads to improved glucose homeostasis in a type 1 diabetic mouse model. Physiol Rep 9: e15007. [Crossref]
- Rahmoune H, Thompson PW, Ward JM, Smith CD, Hong G, et al. (2005) Glucose transporters in human renal proximal tubular cells isolated from the urine of patients with non-insulin-dependent diabetes. Diabetes 54: 3427-3434.
- Ghezzi C, Wright EM (2012) Regulation of the human Na+-dependent glucose cotransporter hSGLT2. Am J Physiol Cell Physiol 303: C348-54. [Crossref]
- Ghezzi C, Loo DDF, Wright EM (2018) Physiology of renal glucose handling via SGLT1, SGLT2 and GLUT2. Diabetologia 61: 2087-2097. [Crossref]
- Santer R, Calado J (2010) Familial renal glucosuria and SGLT2: from a mendelian trait to a therapeutic target. Clin J Am Soc Nephrol 5: 133-141.
- Khoo CM. (2017) Diabetes Mellitus Treatment. In: Quah SR, ed. International Encyclopedia of Public Health (Second Edition). Second Edi. Academic Press 288-293.
- Nair AS, Bagchi D, Lehmann TE, Nair S (2018) Chapter 16 - Renal Sodium-Glucose Transporter-2 Inhibitors as Antidiabetic Agents. In: Bagchi D, Nair S, eds. Nutritional and Therapeutic Interventions for Diabetes and Metabolic Syndrome (Second Edition). Second Edi. Academic Press 207-214.
- Polonsky KS, Burant CF. (2016) Chapter 31 - Type 2 Diabetes Mellitus. In: Melmed S, Polonsky KS, Larsen PR, Kronenberg HM, eds. Williams Textbook of Endocrinology (Thirteenth Edition). Thirteenth. Elsevier 1385-1450.
- Marsenic O (2009) Glucose control by the kidney: an emerging target in diabetes. Am J kidney Dis Off J Natl Kidney Found 53: 875-883. [Crossref]
- De Nicola L, Gabbai FB, Liberti ME, Sagliocca A, Conte G, et al. (2014) Sodium/glucose cotransporter 2 inhibitors and prevention of diabetic nephropathy: targeting the renal tubule in diabetes. Am J kidney Dis Off J Natl Kidney Found 64: 16-24.
- Romere C, Duerrschmid C, Bournat J, et al. (2016) Asprosin, a Fasting-Induced Glucogenic Protein Hormone. Cell 165: 566-579.
- Fahy E, Cotter D, Sud M, Subramaniam S (2011) Lipid classification, structures and tools. Biochim Biophys Acta 1811: 637-647.
- Tabas I (2002) Cholesterol in health and disease. J Clin Invest 110: 583-590. [Crossref]
- Russell DW (1992) Cholesterol biosynthesis and metabolism. Cardiovasc drugs Ther 6: 103-110.
- Dowhan W (1997) The role of phospholipids in cell function. In: Gross RW, ed. Vol 2. Advances in Lipobiology. JAI; 79-107.
- Luo J, Yang H, Song BL. (2020) Mechanisms and regulation of cholesterol homeostasis. Nat Rev Mol Cell Biol 21: 225-245.
- Kirschbaum C, Greis K, Mucha E, et al. (2021) Unravelling the structural complexity of glycolipids with cryogenic infrared spectroscopy. Nat Commun 12: 1201.
- Hanafusa K, Hotta T, Iwabuchi K. (2020) Glycolipids: Linchpins in the Organization and Function of Membrane Microdomains. Front Cell Dev Biol 8. [Crossref]
- Masoro EJ. (1977) Lipids and lipid metabolism. Annu Rev Physiol 39: 301-321.
- Iqbal J, Hussain MM. (2009) Intestinal lipid absorption. Am J Physiol Endocrinol Metab 296: E1183-94.
- Waller DG, Sampson AP (2018) 48 - Lipid disorders. In: Waller DG, Sampson AP, eds. Medical Pharmacology and Therapeutics (Fifth Edition). Fifth Edit. Elsevier 547-557.
- Nguyen P, Leray V, Diez M, et al. (2008) Liver lipid metabolism. J Anim Physiol Anim Nutr 92: 272-283.
- Hewing B, Moore KJ, Fisher EA. (2012) HDL and cardiovascular risk: time to call the plumber? Circ Res 111: 1117-1120. [Crossref]
- Mahdy Ali K, Wonnerth A, Huber K, Wojta J. (2012) Cardiovascular disease risk reduction by raising HDL cholesterol--current therapies and future opportunities. Br J Pharmacol 167: 1177-1194. [Crossref]
- Bhatt S, Kulkarni RN. (2013) Chapter 17 - Significance of Organ Crosstalk in Insulin Resistance and Type 2 Diabetes. In: Karsenty G, ed. Translational Endocrinology of Bone. Academic Press 199-219. [Crossref]
- Fillmore N, Mori J, Lopaschuk GD. (2014) Mitochondrial fatty acid oxidation alterations in heart failure, ischaemic heart disease and diabetic cardiomyopathy. Br J Pharmacol 171: 2080-2090.
- Mihaylova MM, Shaw RJ. (2011) The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol 13: 1016-1023. [Crossref]
- Viollet B, Foretz M, Guigas B, et al. (2006) Activation of AMP-activated protein kinase in the liver: a new strategy for the management of metabolic hepatic disorders. J Physiol 574: 41-53.
- Chen L, Chen XW, Huang X, Song BL, Wang Y, Wang Y (2019) Regulation of glucose and lipid metabolism in health and disease. Sci China Life Sci 62: 1420-1458. [Crossref]
- Yoon M (2009) The role of PPARalpha in lipid metabolism and obesity: focusing on the effects of estrogen on PPARalpha actions. Pharmacol Res 60: 151-159.
- Ponziani FR, Pecere S, Gasbarrini A, Ojetti V (2015) Physiology and pathophysiology of liver lipid metabolism. Expert Rev Gastroenterol Hepatol 9: 1055-1067. [Crossref]
- Souza-Mello V. (2015) Peroxisome proliferator-activated receptors as targets to treat non-alcoholic fatty liver disease. World J Hepatol 7:1012-1019.
- Koba S, Hirano T. (2011) [Dyslipidemia and atherosclerosis]. Nihon Rinsho 69: 138-143.
- Byrne CD, Targher G. (2015) NAFLD: a multisystem disease. J Hepatol 62: S47-64.
- Kořínková L, Pražienková V, Černá L, et al. (2020) Pathophysiology of NAFLD and NASH in Experimental Models: The Role of Food Intake Regulating Peptides. Front Endocrinol (Lausanne) 11: 597583.
- Swain M, Nath P, Parida PK, et al. (2017) Biochemical Profile of Nonalcoholic Fatty Liver Disease Patients in Eastern India with Histopathological Correlation. Indian J Clin Biochem 32: 306-314. [Crossref]
- Cohen DE, Fisher EA. (2013) Lipoprotein metabolism, dyslipidemia, and nonalcoholic fatty liver disease. Semin Liver Dis 33: 380-388. [Crossref]
- Pei K, Gui T, Kan D, et al. (2020) An Overview of Lipid Metabolism and Nonalcoholic Fatty Liver Disease. Biomed Res Int 2020: 4020249. [Crossref]
- Okada Y, Yamaguchi K, Nakajima T, et al. (2013) Rosuvastatin ameliorates high-fat and high-cholesterol diet-induced nonalcoholic steatohepatitis in rats. Liver Int Off J Int Assoc Study Liver 33: 301-311. [Crossref]
- Ji G, Zhao X, Leng L, Liu P, Jiang Z. (2011) Comparison of dietary control and atorvastatin on high fat diet induced hepatic steatosis and hyperlipidemia in rats. Lipids Health Dis 10: 23. [Crossref]
- Chong LW, Hsu YC, Lee TF, et al. (2015) Fluvastatin attenuates hepatic steatosis-induced fibrogenesis in rats through inhibiting paracrine effect of hepatocyte on hepatic stellate cells. BMC Gastroenterol 15: 22.
- Athyros VG, Mikhailidis DP, Didangelos TP, et al. (2006) Effect of multifactorial treatment on non-alcoholic fatty liver disease in metabolic syndrome: a randomised study. Curr Med Res Opin 22: 873-883. [Crossref]
- Lonardo A, Sookoian S, Pirola CJ, Targher G. (2016) Non-alcoholic fatty liver disease and risk of cardiovascular disease. Metabolism 65:1136-1150.
- Hyogo H, Tazuma S, Arihiro K, et al. (2008) Efficacy of atorvastatin for the treatment of nonalcoholic steatohepatitis with dyslipidemia. Metabolism 57: 1711-1718. [Crossref]
- Singh S, Singh PP, Singh AG, Murad MH, Sanchez W. (2013) Statins are associated with a reduced risk of hepatocellular cancer: a systematic review and meta-analysis. Gastroenterology 144: 323-332.
- Ratziu V, Harrison SA, Francque S, et al. (2016) Elafibranor, an Agonist of the Peroxisome Proliferator-Activated Receptor-α and -δ, Induces Resolution of Nonalcoholic Steatohepatitis Without Fibrosis Worsening. Gastroenterology 150: 1147-1159.e5.
- Pawlak M, Lefebvre P, Staels B (2015) Molecular mechanism of PPARα action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J Hepatol 62: 720-733. [Crossref]
- Fiorucci S, Distrutti E, Carino A, Zampella A, Biagioli M, et al. (2021) Bile acids and their receptors in metabolic disorders. Prog Lipid Res 82: 101094.
- Ni M, Zhang B, Zhao J, et al. (2019) Biological mechanisms and related natural modulators of liver X receptor in nonalcoholic fatty liver disease. Biomed Pharmacother 113: 108778.
- Akerib DS, Alsum S, Aquino C, et al. (2012) A role for proteinase-activated receptor-1 in inflammatory bowel diseases. Gastroenterology 10: 21303.
- Wang M, Sun S, Wu T, et al. (2013) Inhibition of LXRα/SREBP-1c-Mediated Hepatic Steatosis by Jiang-Zhi Granule. Evid Based Complement Alternat Med 2013: 584634. [Crossref]
- Kurtz R, Anderman MF, Shepard BD. (2021) GPCRs get fatty: the role of G protein-coupled receptor signaling in the development and progression of nonalcoholic fatty liver disease. Am J Physiol Gastrointest Liver Physiol 320: G304-G318.
- Zhang S, Li L, Li H. (2021) Role of ectopic olfactory receptors in glucose and lipid metabolism. Br J Pharmacol 178: 4792-4807.
- Hong JY, Lee BH, Kim TH, et al. (2013) GC-MS/MS method for the quantification of α-cedrene in rat plasma and its pharmacokinetic application. J Sep Sci 36: 3558-3562.
- Iida T, Ubukata M, Mitani I, et al. (2018) Discovery of potent liver-selective stearoyl-CoA desaturase-1 (SCD1) inhibitors, thiazole-4-acetic acid derivatives, for the treatment of diabetes, hepatic steatosis, and obesity. Eur J Med Chem 158: 832-852. [Crossref]
- Lindén D, William-Olsson L, Rhedin M, Asztély AK, Clapham JC, et al. (2004) Overexpression of mitochondrial GPAT in rat hepatocytes leads to decreased fatty acid oxidation and increased glycerolipid biosynthesis. J Lipid Res 45: 1279-1288. [Crossref]
- Himms-Hagen J. (1989) Role of thermogenesis in the regulation of energy balance in relation to obesity. Can J Physiol Pharmacol 67: 394-401.
- Alcalá M, Calderon-Dominguez M, Serra D, Herrero L, Viana M, et al. (2019) Mechanisms of Impaired Brown Adipose Tissue Recruitment in Obesity. Front Physiol 10.
- Cypess AM, Kahn CR (2010) Brown fat as a therapy for obesity and diabetes. Curr Opin Endocrinol Diabetes Obes 17: 143-149.
- Knebel B, Hartwig S, Jacob S, et al. (2018) Inactivation of SREBP-1a Phosphorylation Prevents Fatty Liver Disease in Mice: Identification of Related Signaling Pathways by Gene Expression Profiles in Liver and Proteomes of Peroxisomes. Int J Mol Sci 19(4). [Crossref]
- Eberlé D, Hegarty B, Bossard P, Ferré P, Foufelle F, et al. (2004) SREBP transcription factors: master regulators of lipid homeostasis. Biochimie 86: 839-848. [Crossref]
- Grosmaitre X, Vassalli A, Mombaerts P, Shepherd GM, Ma M, et al. (2006) Odorant responses of olfactory sensory neurons expressing the odorant receptor MOR23: a patch clamp analysis in gene-targeted mice. Proc Natl Acad Sci U S A 103: 1970-1975.
- Mehmood S, Orhan I, Ahsan Z, Aslan S, Gulfraz M, et al. (2008) Fatty acid composition of seed oil of different Sorghum bicolor varieties. Food Chem 109: 855-859. [Crossref]
- Muthulakshmi S, Saravanan R. (2013) Protective effects of azelaic acid against high-fat diet-induced oxidative stress in liver, kidney and heart of C57BL/6J mice. Mol Cell Biochem 377: 23-33. [Crossref]
- Muthulakshmi S, Chakrabarti AK, Mukherjee S. (2015) Gene expression profile of high-fat diet-fed C57BL/6J mice: in search of potential role of azelaic acid. J Physiol Biochem 71: 29-42.
- Abaffy T, Matsunami H, Luetje CW. (2006) Functional analysis of a mammalian odorant receptor subfamily. J Neurochem 97: 1506-1518. [Crossref]
- Morcia C, Tumino G, Ghizzoni R, Terzi V. (2016) Carvone (Mentha spicata L.) Oils 309-316.
- Bushdid C, de March CA, Fiorucci S, Matsunami H, Golebiowski J, et al. (2018) Agonists of G-Protein-Coupled Odorant Receptors Are Predicted from Chemical Features. J Phys Chem Lett 9: 2235-2240.
- Diels S, Huybreghts S, Van Hoorenbeeck K, et al. (2020) Copy number variant analysis and expression profiling of the olfactory receptor-rich 11q11 region in obesity predisposition. Mol Genet Metab Reports 25: 100656. [Crossref]
- Giusepponi ME, Kern M, Chakaroun R, et al. (2018) Gene expression profiling in adipose tissue of Sprague Dawley rats identifies olfactory receptor 984 as a potential obesity treatment target. Biochem Biophys Res Commun 505: 801-806.
- Jarick I, Vogel CIG, Scherag S, et al. (2010) Novel common copy number variation for early onset extreme obesity on chromosome 11q11 identified by a genome-wide analysis. Hum Mol Genet 20: 840-852. [Crossref]
- Dharmalingam M, Yamasandhi PG. (2018) Nonalcoholic Fatty Liver Disease and Type 2 Diabetes Mellitus. Indian J Endocrinol Metab 22: 421-428.
- Asrih M, Jornayvaz FR. (2015) Metabolic syndrome and nonalcoholic fatty liver disease: Is insulin resistance the link? Mol Cell Endocrinol 418: 55-65. [Crossref]
- Zieve FJ. (2004) The metabolic syndrome: diagnosis and treatment. Clin Cornerstone 6 Suppl 3: S5-13.
- Faour M, Magnan C, Gurden H, Martin C. (2022) Olfaction in the context of obesity and diabetes: Insights from animal models to humans. Neuropharmacology 206: 108923. [Crossref]
- Perlman RL. (2016) Mouse models of human disease: An evolutionary perspective. Evol Med public Heal 2016: 170-176. [Crossref]



 Azelaic acid
Azelaic acid Azelaic acid
Azelaic acid Octanoic acid
Octanoic acid Sebacic acid
Sebacic acid Undecanoic acid
Undecanoic acid

 Citronellal
Citronellal (-)-Carvone
(-)-Carvone Citral
Citral Nerol
Nerol
 Nonanoic acid
Nonanoic acid α-cedrene
α-cedrene Octanoic acid
Octanoic acid Cyclohexanone
Cyclohexanone Cycloheptanone
Cycloheptanone
 Cyclooctanone
Cyclooctanone
