Reconstitution of cell lines and occurrence of complications following hematopoietic stem cell transplantation (HSCT) are regulated by genome expression. Microarray technique allows for simultaneous assessment of expression of nearly all human genes. The objective of the study was to compare whole genome expression in children before and after HSCT.
A total of 44 children treated with HSCT were enrolled in the study. Gene expression was measured before HSCT (pre-HSCT group; n=44) and after a median of 6 months after allogenic HSCT (post-HSCT group; n=27; all children were included in the pre-HSCT group). Neoplasms were the indication for HSCT in 73% of the patients. Whole genome expression was assessed in leukocytes using GeneChip® HumanGene 1.0 ST microarray.
The GeneCards Human Genes Database was used for analysis of genes function. The analysis of genomic profiles before and after HSCT revealed 18 significantly different genes with defined function. These genes are responsible for proliferation and differentiation of cells (14 genes), apoptosis (8 genes), migration of cells (3 genes) and fluid/electrolyte homeostasis (2 genes).
Conclusions: Activation of genes involved in reconstitution of donor cell lines, and those related to immune reactions observed after HSCT, form the genetic background for physiological and pathological processes following HSCT.
stem cell transplantation, immune reactions, microarrays, children
Allogeneic hematopoietic stem cell transplantation (HSCT) is currently a safe and life-saving treatment modality used in a wide spectrum of diseases [1-3].
However, transplant recipients are still at high risk of morbidity and mortality due to delay of reconstitution of cell lineages, prolonged treatment with multiple, nonspecific and toxic immunosuppressive drugs, and immune reactions like graft versus host disease (GvHD) or graft rejection (GR) [4-8]. All the observed processes are regulated by genetic machinery [9-11].
Two recent articles explain the role of gene analyses in the search for immunotherapy targets in malignant diseases and in the study of immune complications after HSCT. In the study by Pont MJ et al. microarray gene expression analysis on malignant and healthy hematopoietic cells and various non-hematopoietic cell types cultured under steady state and inflammatory conditions was performed. The goal of the research was to evaluate the value of the microarray dataset to analyze and select potential targets for immunotherapy of hematological malignancies. Gene expression profiles were generated for hematopoiesis-restricted minor histocompatibility antigens that are recognized by specific T cells in the context of HLA. The analysis confirmed restricted or predominant expression of these antigens in hematopoietic cells, but ARHGDIB and ITGB2 also showed intermediate and low expression in endothelial cells and fibroblasts, respectively. In addition to minor histocompatibility antigens, the authors generated gene expression profiles for surface antigens with known B-cell specific expression. Microarray analysis confirmed restricted or predominant expression of these antigens in (malignant) B cells. However, CD79B was also expressed in a variety of other (non-)hematopoietic cell types. For ROR1, microarray data confirmed overexpression in CLL as compared to healthy B cells. ROR1 expression was also found in biliary epithelialcells and to variable extents in skin fibroblasts. Those cell types originate from organs that are often targeted in detrimental immune responses after allogeneic stem cell transplantation leading to graft versus-host disease [12].
In the second study by Inamoto Y et al. [13] candidate single-nucleotide polymorphisms (SNPs)were examined that have a well-documented association with systemic sclerosis to determine whether these SNPs are also associated with the risk of sclerotic GVHD. The study cohort included 847 consecutive patients who were diagnosed with chronic GVHD. Genotyping was performed using microarrays, followed by imputation of unobserved SNPs. The donor rs10516487 TT genotype was associated with lower risk of sclerotic GVHD. Donor and recipient rs2056626 GG or GT genotypes and rs987870 CC genotypes were associated with higher risk of sclerotic GVHD. Additionally, the recipient DPA1*01:03∼DPB1*04:01 haplotype and certain amino acid substitutions in the recipient P1 peptide-binding pocket of the HLA DP heterodimer were associated with risk of sclerotic GVHD. Genetic components associated with systemic sclerosis are also associated with sclerotic GVHD. Authors concluded that HLA-DP–mediated antigen presentation, T-cell response, and B-cell activation have important roles in the pathogenic mechanisms of both diseases [13].
The aim of our study was to analyze the genome expression changes resulting from HSCT in pediatric population.
This was a prospective study, the study group consisted of children and teenagers referred for HSCT, which was performed according to disease-specific treatment protocols. The study was approved by the Ethics Committee of the Jagiellonian University (KBET/96/B/2008). Written informed consent was obtained from all parents and from all patients ≥16 years of age.
Microarray analysis
Blood samples (1.5 mL) were collected from each patient before conditioning and approximately six months after HSCT (median 6.3 months). Total RNA extraction from blood mononuclears was performed using RiboPure Blood Kit (Ambion, Life Technologies, Carlsbad, USA). Whole genome expression was assessed using GeneChip Human Gene 1.0 ST Arrays (Affymetrix, Santa Clara, USA). Whole transcript microarray experiment was performed according to the manufacturer’s protocol (GeneChip Whole Transcript sense Target Labeling Assay Manual, Version 4).
Statistical analysis
The microarray data were preprocessed using the R/Bioconductor package [14-16]. Robust Multiarray Average (RMA) was used for normalization [17]. Quality control was performed by investigating Principal Component Analysis (PCA), Relative Log Expression (RLE) and Normalized Unscaled Standard Error (NUSE) plots.
Moderated t-tests [18] were used to detect the probes with different expression in various groups; the limma package [19] from the R software was used. It was assumed that the log2 transformed gene expression levels are normally distributed and the between-group variation is of comparable magnitude. Multiple testing correction (Benjamini-Hochberg procedure) was applied to control the false discovery rate (FDR) [20]. Significantly different expression in the probe sets was defined as multiple comparison-corrected two-sided p-value <0.05. For comparison of gene expression levels (33000 genes studied) between the group of patients assessed before HSCT (pre-HSCT group) and the group of patients assessed after HSCT (post-HSCT group) ANOVA filtering by p-value 0.00005 was used with expected false positive results equal to 2.
The GeneCards Human Genes Database (www.genecards.org/) was used for analysis of genes function.
The study groups
The pre-HSCT group included 44 patients (31 boys, 13 girls) aged 1.5 – 19 (median 10.5) years, referred to the Stem Cell Transplantation Centre of the University Children’s Hospital in Krakow. Neoplasms were the indication for HSCT in 73% of children. The indications for HSCT are listed in Table 1 and types of HSCT procedures are listed in Table 2.
Table 1. Indications for HSCT (pre-HSCT group)
Diagnosis |
N ( %) |
Acute lymphoblastic leukemia (ALL) |
17 (38.6) |
Acute myeloblastic leukemia (AML) |
5 (11.3) |
Acute bilineage leukemia (ABL) |
1 (2.3) |
Chronic myelocytic leukemia (CML) |
1 (2.3) |
Hodgkin Lymphoma (HD) |
2 (4.6) |
Myelodysplastic syndrome (MDS) |
1 (2.3) |
Juvenile myelomonocytic leukemia (JMML) and AML |
1 (2.3) |
Neuroblastoma (NBL) |
4 (9) |
Neoplastic diseases–total |
32 (73) |
Severe aplastic anemia (SAA) |
4 (9) |
Blackfan-Diamond Anemia (BDA) |
1 (2.3) |
Fanconi Anemia (FA) |
1 (2.3) |
Chronic granulomatous disease (CGD) |
3 (6.8) |
Autoimmune lymphoproliferative syndrome (ALPS) |
1 (2.3) |
Hyper IgM syndrome (HIgM) |
1 (2.3) |
Inherited neutropenia (IN) |
1 (2.3) |
Non-neoplastic diseases–total |
12 (27) |
Table 2. Types of HSCT procedures (ALL: Acute lymphoblastic leukemia; ALPS: Autoimmune lymphoproliferative syndrome; AML: Acute myeloblastic leukemia; CGD: Chronic granulomatous disease; HIgM–hyper IgM syndrome; JMML: Juvenile myelomonocytic leukemia; MDS: Myelodysplastic syndrome; SAA: Severe aplastic anemia)
Type of HSCT |
n(%) |
Disease (n) |
Allogeneic
n=27 (100%) |
MUD-16 (59)
|
ALL-9
AML-4
SAA-1
CGD–2 |
MSD-9 (33)
|
ALL-3
SAA-2
JMML and AML-1
CGD-1
HIgM-1
MDS–1 |
MFD-2 (7) |
SAA-1
ALPS–1 |
The post-HSCT group included 27 patients from the pre-HSCT group, aged 2.8 – 19.5 (average 10.4) years. Six patients from the pre-HSCT group were excluded from the analysis due to technical reasons (poor quality of RNA sample) and another eleven patients were lost to follow up. Four children in the post-HSCT group died due to complications of treatment or disease progression, one child died because of GvHD. All patients in the post-HSCT group were treated with ablative conditioning regimens (Table 2). The key clinical characteristics of the patients are presented in Table 3.
Table 3. Characteristics of the study groups (GvHD–graft-versus-host disease)
Pre-HSCT group |
N |
44 |
Sex |
Boys 31, Girls 13 |
Age (years) |
1.5–19 (median 10.5) |
Chemotherapy before HSCT (%)
First line
Second line
>third line |
23 (71%)
13
9
1 |
Local radiotherapy
Cranial (dose)
Testes |
7
5 (12Gy-4, 18Gy-1)
2 (12Gy/24Gy, 18Gy/18Gy) |
Time since diagnosis and patient selection (years) |
Median, 1.4; range 0.08 –12.9 |
Post-HSCT group |
N |
27 |
Sex |
Boys 20, Girls 7 |
Age (years) |
2.8– 19.5 (median 9.9) |
Conditioning regimen based on busulfan/treosulfan (n) |
16 (59%) |
Total body irradiation–12Gy/6 fractions (n) |
7 (26%) |
GvHD prophylaxis (n)
Cyclosporin
Methotrexate+cyclosporin |
4 (15%)
23 (85%) |
GvHD (n) |
14(52%) |
Median time from HSCT to the second assessment (range) |
6.3 (5.9-19.1) months |
Systemic glucocorticoids (%) |
25 (93%) |
Median and range of cumulative doses of glucocorticoids (equivalent of prednisone) |
1531 (29–9758) mg/m2 |
Median duration of systemic glucocorticoid therapy |
73 (1–315) days |
Median time since discontinuation of glucocorticoids (range) |
3.5 (0–14.3) months |
Median time from discontinuation of immunosuppressive treatment to the second assessment (range) (16 patients) |
2 (0–9.5) months |
Whole genome expression
All the primary microarray data were submitted to GEO public repository and are accessible using GEO Series accession number GSE88852 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE88852).
A summary of the differentially expressed genes and their function is presented in Table 4 and Figure 1.
Table 4. The differentially expressed genes and their function(www.genecards.org/)
Gene |
Function of coded protein |
Final effect |
Activation of function in post-HSCT group |
WBP1L |
Expressed in hematopoietic tissues
Proapoptotic
Predispostion to acute leukemia |
Activation of proliferation and apoptosis |
U2AF1 |
Hematopoiesis regulator |
Regulation of hematopoiesis |
AREL1 |
Apoptosis inhibitor |
Inhibition of apoptosis |
Lasp-1 |
Interactions with other proteins, carries signals in cytoplasm and nuclei
Key role in cell structure, physiologic and signaling processes
Overexpression influence cancer progression |
Activation of proliferation |
IQSEC1 |
|
Cells migration |
CSTN3 |
- Modulator of calcium-mediated postsynaptic signals
- Modulator of endocrine function
- Acceleration of neuronal death
- Upregulation in cortical neurons increase vulnerability of neurons
|
Regulation of signalization
Regulation of endocrine function
Apoptosis activation |
ZFP319 |
Implicated in regulating differentiation and cell fate determination
|
Regulation of differentiation Activation of apoptosis |
CST7 |
Induction of proliferation
Immune regulation through inhibition of a unique target in the hematopoietic system
Expression is correlated with metastatic potential of malignant tumors |
Activation of proliferation
Regulation of the hematopoietic system |
EDARADD |
Encodes a member of the TNF receptor family
Activation of cell death pathways
Development of ectodermal derivatives |
Activation of proliferation
Activation of apoptosis |
ECOP |
Stimulation of proliferation of neoplastic cells Inhibition of apoptosis
Signal transducer activity
Control of the intracellular redox state |
Activation of proliferation
Inhibition of apoptosis |
FAM53B |
Activation of cells proliferation, maintenance of pluripotent status |
Activation of proliferation |
Inhibition of function in post-HSCT group |
B3GALT2 |
Embryogenesis regulator |
Inhibition of proliferation |
SLC4A10 |
- Regulation of intracellular pH (brain, kidney, small intestine)
|
Regulation of fluid end electrolyte homeostasis |
TRMT5 |
Important role in hematopoiesis
Regulation of polypeptide synthesis |
Inhibition of hematopoiesis |
NR3C2 |
Encodes mineralocorticoid receptor
Regulation of aldosterone effects on water and electrolyte homeostasis
Regulation of T-cell migration and redistribution of T-cell subsets to lymph nodes |
Regulation of fluid and electrolyte homeostasis
Inhibition of T-cell migration |
PLAG1 |
Regulation of DNA and RNA transcription |
Inhibition of proliferation |
DPP IV |
- Activation of T-cell receptor-mediated T-cell activation
- Induces T-cell proliferation by binding to chemokines, mitogenic growth factors , neuropeptides, and peptide hormones
- Promotes lymphocyte-epithelial cell adhesion, migration, tube formation
- Regulation of expression of hemoglobin genes
|
Inhibition of T-cells function
Inhibition of proliferation
Inhibition of erythropoiesis |
ST13 |
Suppressor gene
Inhibition of growth and migration of neoplastic cells |
Activation of proliferation and migration |
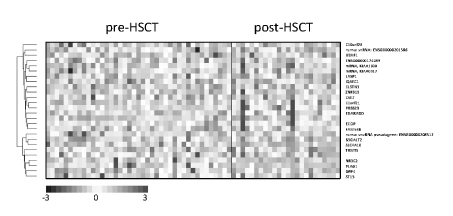
Figure 1. Clustering of signals of the assessed genes.
A comparison of the pre-HSCT and post-HSCT groups revealed 26 genes having the highest expression differences (p-value 0.00005, expected false positive results equal to 2)–Figure 1. Based on search in the GeneCards - Human Genes Database (www.genecards.org/) well defined function was found in 18 cases. In the post-HSCT group gene activation (14 genes) was seen a little more frequently than gene inhibition (10 genes). Proliferation-related genes were activated (WBP1L, Lasp-1, CST7, EDARADD, ECOP, FAM53B, ST13) or suppressed (B3GALT2, TRMT5, PLAG1, DPPIV). Similarly, several apoptosis-related genes were activated (WBP1L, CSTN3, ZFP319, CST7, EDARADD) or suppressed (AREL1, ECOP, B3GALT2). Three genes were responsible for regulation of migration of cells, including lymphocytes T (IQSEC1, ST13, NR3C2). The function of T-cells was inhibited by DPPIV. Increased expression was observed for CST7. This gene is responsible for immune regulation through inhibition of a unique target in the hematopoietic system. Increased expression was also detected for genes ZFP319 and U2AF1 regulating differentiation and hematopoiesis respectively. On the other hand, inhibition of hematopoiesis was represented by TRMT5 and inhibition of erythropoiesis by DPPIV genes. Moreover, a decrease in expression of SLC4A10 and NR3C2 – genes regulating fluid and electrolyte homeostasis - was found.
In our earlier study including 27 children treated with HSCT, we used pathway enrichment analysis of 250 overexpressed genes, and we identified five genomic pathways: “Allograft rejection”, “Graft-versus-host disease”, “Type I diabetes mellitus”, “Autoimmune thyroid disease” and “Viral myocarditis” [21]. In the present study we compared genome expression between the groups of children before HSCT and after allogeneic HSCT. Using conservative statistical criteria (p-value 0.00005 with expected false positive results equal to 2) a total of 24 genes with well-defined function were identified among approximately 33000 genes. Thus, whole genome expression of autologous lymphocytes of 44 children treated with HSCT was compared with genome expression of lymphocytes of 27 allogeneic donors in the recipient’s microenvironment. The lymphocytes play basic roles in all immune reactions after HSCT.
Consequently, activation of specific genes in allogeneic lymphocytes genomes is responsible for regulation of engraftment of hematopoietic system, immune reconstitution and immune complication seen after HSCT [13]. In our study gene activation (14 genes) was seen a little more frequently than gene inhibition (10 genes). The balance between proliferation/migration providing to engraftment/reconstitution of cell lines and apoptosis represent the key processes of correction of cells population size [1,13]. Consequently, GvHD was the key factor responsible for outcomes of HSCT. The observed inhibition of genes regulating fluid and electrolyte homeostasis is understandable as volume overload and electrolyte disturbances are common complications of HSCT. Erythrocytosis is observed in approximately 36% of children after several years of follow up (unpublished data). Thus, inhibition of donor genes regulating hematopoiesis and erythropoiesis, which undergo stimulation by recipient’s tissues, previously chronically hypoxic due to prolonged anemia, might be a protective mechanism. Four (15%) patients relapsed after HSCT. As some of the genes stimulating proliferation were described as being overexpressed in various types of cancers(www.genecards.org/), it is possible that they took part in propagation of relapses in our patients.
In conclusion, the observed activation of genes involved in reconstitution of donor cell lines and those related to immune reactions observed after HSCT seem to form a genetic background of the common processes and immune complications of HSCT. Our data correspond with our previous results indicating pathways of allogenic reactions of donor cells against recipient tissues.
The authors declare no conflict of interest.
This work was supported by national grant number NN 407 198737
KK, SS designed and performed research, analyzed and interpreted data, and wrote the manuscript. MBM analyzed and interpreted data and helped in drafting the manuscript. AG, MK, MP and KF performed research and collected data. WS, DP analyzed and interpreted data. WB conducted the clinical protocols and interpreted data.
- Ferrara JL, Levine JE, Reddy P, Holler E (2009) Graft-versus-host disease. Lancet 373: 1550-1561. [Crossref]
- Engel KL, Mackiewicz M, Hardigan AA, Myers RM, Savic D (2016) Decoding transcriptional enhancers: Evolving from annotation to functional interpretation. Semin Cell Dev Biol S1084-9521: 30140-30149.
- Pont MJ, Honders MW, Kremer AN, van Kooten C, Out C, et al. (2016) Microarray gene expression analysis to evaluate cell type specific expression of targets relevant for immunotherapy of hematological malignancies. PLoS One 11: e0155165.
- Inamoto Y, Martin PJ, Flowers ME, Lee SJ, Carpenter PA, et al. (2016) Genetic risk factors for sclerotic graft-versus-host disease. Blood 128: 1516-1524. [Crossref]
- Lee MT (2004) Power and sample size considerations. In: Analysis of microarray expression Data, 1st ed. Boston: Kluwer academic publishers.
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80.
- Bengtsson H, Simpson K, Bullard J, Hansen K (2004) Aroma.affymetrix: A generic framework in R for analyzing small to very large Affymetrix data sets in bounded memory. Tech Rep #745, Department of statistics, University of California, Berkeley, USA.
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, et al. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249-264.
- Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: 1-25.
- Smyth GK (2005) Linear Models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and computational biology solutions using R and bioconductor. New York: Springer Verlag.
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B Stat Methodol 57: 289-300.
- Skoczen S, Bik-Multanowski M, Pietrzyk JJ, Grabowska A, Fijorek K, at al. (2016) Genetic background of immune complications after allogeneic hematopoietic stem cell transplantation in children. Stem Cells Int.
- Flowers ME, Martin PJ (2015) How we treat chronic graft-versus-host disease. Blood 125: 606-615. [Crossref]
- Socie G, Ritz J (2014) Current issues in chronic graft-versus-host disease. Blood 124: 374-384. [Crossref]
- Nikiforow S, Alyea EP (2014) Maximizing GVL in allogeneic transplantation: Role of donor lymphocyte infusions. Hematology Am Soc Hematol Educ Program 5: 570-575.
- Strober S (2014) Path to clinical transplantation tolerance and prevention of graft-versus-host disease. Immunol Res 58: 240-248. [Crossref]
- Choi SW, Reddy P (2014) Current and emerging strategies for the prevention of graft-versus-host disease. Nat Rev Clin Oncol 11: 536-547. [Crossref]
- Paczesny S (2013) Discovery and validation of graft-versus-host disease biomarkers. Blood 121: 585-594. [Crossref]
- Heidegger S, van den Brink MR, Haas T, Poeck H (2014) The role of pattern recognition receptors in graft-versus-host disease and graft-versus-leukemia after allogeneic stem cell transplantation. Front Immunol 18: 1-7.
- Markey KA, MacDonald KP, Hill GR (2014) The biology of graft-versus host disease: experimental systems instructing clinical practice. Blood 124: 354-362.
- Reikvam H, Gronningsæter IS, Ahmed AB, Hatfield K, Bruserud o (2015) Metabolic serum profiles for patients receiving allogeneic stem cell transplantation: The pretransplant profile differs for patients with and without posttransplant capillary leak syndrome. Dis Markers.

