Abstract
Waldenström's macroglobulinemia (WM) is categorized as a B-cell neoplasia characterized by lymphoplasmacytic infiltration in the bone marrow and the overproduction of serum monoclonal IgM. The precise cause of WM is unknown, but familial patterns suggest the possibility of an underlying genetic defect. The purpose of this study was to determine the association between different gene variations and the development of WM. We studied different genetic variations reporting significant incidence rates. The most representative of those were the MYD88 (L256P) mutation with an incidence rate of 81% and 6q deletion with an incidence rate of 36%. In addition, CXCR4 mutation, 13q14 deletion and trisomy 4 showed an incidence rate of 29%, 12% and 14%, respectively. Finally, the deletion 17p13 was present in 8% of subjects. Our results suggest that the MYD88 (L256P) mutation might be a target for the development of specific therapeutic strategies.
Keywords
Waldenström's macroglobulinemia, genetics, genes, DNA mutation, hematology
Introduction
Waldenström's macroglobulinemia (WM) is a B-cell neoplasia characterized by lymphoplasmacytic infiltration in the bone marrow and overproduction of monoclonal IgM. WM is considered a rare form of neoplasia, accounting for only 1−2% of all hematological tumors [1], with an incidence of 3−4 cases per million people per year [2]. WM remains an incurable disease [3]. Current available chemotherapeutic options include alkylating agents, purine analogs, nucleosides and rituximab, a monoclonal antibody that binds specifically to the CD20 B-cell antigen [4]. The diagnosis is based on a serum monoclonal IgM component associated with lymphoplasmacytic infiltration in the bone marrow irrespective of the presence or absence of symptoms [5]. Although WM is considered a separate pathological clinical entity, the absence of morphologic, immunophenotypic or specific chromosome markers makes it difficult to distinguish this condition from other B-cell neoplasias [6]. A level of monoclonal IgM > 1.5 g·dl-1 is characteristic of WM and is used to differentiate WM from other forms of neoplasia with plasmacytoid differentiation.
The etiology of WM remains largely unknown, although aging is an important factor (the average age of diagnosis is ~70 years) [7,8] as well as sex and ethnicity (the incidence is 2 times higher in men than in women and 3.2 times higher in white than in black people [9]). Since the first report describing familial WM in 1962, various cases, case-control and cohort studies have been published showing indications of a strong hereditary predisposition to WM [4,10]. Emerging population data and clinical studies have confirmed an increased risk for WM patients’ family members who are diagnosed with WM or a related B-cell disorder. Moreover, the spectrum of familial WM can include relatives with IgM monoclonal gammopathy and/or immunological disorders [11]. Several familial aggregation studies of WM implicate a role both for environmental and hereditary factors [11]. Also, several reports suggest that genetic mutations or genetic polymorphisms can be associated with the risk of WM. The evidence of somatic mutations of the immunoglobulin gene in WM indicates a role for antigenic stimulation in the development of WM [10]. Therefore, the objective of this study was to examine the evidence for an association between the different genetic variations and the development of WM.
Sources
Electronic literature searches were performed to identify all scientific and clinical reports published regarding WM and its genetic associations. The search was conducted in three different databases, Scopus, Science Direct and PubMed, from their inception to May 2017 using the search terms “Waldenström macroglobulinemia”, “genetic factors”, “gene”, “genetic”, “genetics”, “Waldenström´s macroglobulinemia” and “whole genome sequencing”, as well as the combination of one or more of these terms. A total number of 10,704 articles were identified (1,278 in PubMed, 758 in Scopus and 8,668 in Science Direct).
The inclusion criteria for this analysis were the following: i) studies with WM patients; ii) genetic studies and studies on single nucleotide polymorphisms (SNPs); and iii) retrospective observational studies. The exclusion criteria were: i) articles of systematic revision or meta-analysis; ii) conference proceedings; and iii) case studies.
We followed the recommendations of the PRISMA statement [12]. From the 10,704 articles identified during the bibliographic search, the established criteria of inclusion were applied to determine potential articles. From each selected article, the number of study subjects was collected, as well as the characteristics of each individual (age, age range, sex, and race), the gene or genes investigated in each publication and the results of each of the variables subjected to study. Two authors independently assessed the obtained records by the search results.
A meta-analysis of percentages using a model of variable effects was performed to estimate the combined incidence rate of the patients who presented with deletion of chromosome 6q, the L256P mutation in the myeloid differentiation primary response (MYD88) gene, CXCR4 mutation, deletion of chromosome 13q14, trisomy of chromosome 4 and deletion of chromosome 17p13. To assign the importance of studies in this type of meta-analysis, it is not recommended to calculate the standardized error rate given that it can deliver some imprecise confidence intervals [13]. Instead, the importance of the studies was determined by the sample size and the standard error of RR transformed by the Freeman-Tukey double arcsine. Egger’s regression test was used to assess evidence of publication bias. The presence of heterogeneity between the studies was assessed by Cochran’s Q test and the I2 index. The level of significance was determined as p ≤ 0.05. All statistical analysis was carried out using MIX 2.0 Pro for Excel.
Results
From an initial identification of 10,704 studies, 10,552 were excluded according to the above-mentioned criteria. From the remaining 152 potentially relevant studies, 34 studies were considered relevant and of these 29 were included in the meta-analysis. The flow chart (Figure 1) provides an overview of the biographical search and the included and excluded data. A summary of key data and genetic variables from the 27 relevant articles is presented in Table 1. Data such as average age and sex were also included, with the latter expressed as the percentage of women who participated in the study.
Table 1. Genetic variables in Waldenström's macroglobulinemia
Author |
n |
% Women
Average age |
|
Variable studied |
Results |
Included in the meta-analysis |
Ansell SM, et al. [23] |
39 |
|
|
Mutation MYD88 (L265P) |
96.6% |
✓ |
Ada I Rivera [24] |
1 |
|
77 |
Trisomy 4 |
100% |
x |
Ballester LY, et al [25] |
32
33 |
|
|
CXCR4
Mutation MYD88 (L265P)
|
25%
69.69%
|
✓
✓ |
Braggio E, et al. [26] |
42 |
|
|
Deletion 6q |
40% |
✓ |
Gain 6p |
17% |
x |
Trisomy 3 |
20-50% |
x |
Gain 3q13-q28 |
10% |
x |
Gain 18q |
17% |
x |
Gain 4q |
12% |
x |
Gain 8q |
10% |
x |
Braggio E, et al. [27] |
42 |
|
|
Deletion 6q |
43% |
✓ |
Trisomy 4 |
12% |
✓ |
Deletion 17p13 |
10% |
✓ |
Braggio E, et al. [6] |
42 |
|
|
Gain 6p |
17% |
x |
Deletion 13q14 |
10% |
✓ |
Deletion 7q22 |
7.10% |
x |
Deletion 8p |
7.10% |
x |
Deletion 11q22-23 |
7.10% |
x |
Deletion 11q23.2-24 |
7.10% |
x |
Deletion 17p11.2-p13.3 |
7.10% |
x |
Trisomy 18 |
16.60% |
x |
Gain 6p |
16.60% |
x |
Trisomy 4 |
19% |
✓ |
Trisomy 3 |
9.50% |
x |
Trisomy 8q |
9.50% |
x |
Xq27-q28 |
9.50% |
x |
Chang HQ et al. [28] |
77 |
|
|
Deletion 6q |
41.60% |
✓ |
Chang H et al. [29] |
34 |
|
|
Deletion 6q |
38% |
✓ |
Ghobrial IM, et al [30] |
20 |
|
|
CXCR4 (C1013G)
Mutation MYD88 L265P |
5%
80% |
✓
✓ |
Hunter Z, et al. [22] |
30 |
|
|
Mutation MYD88 L265P |
90% |
✓ |
Deletion 6q
CXCR4 |
43%
27% |
✓ |
Trisomy 4 |
23% |
✓ |
Jimenez C [31] |
117 |
|
|
Mutation MYD88 L265P |
86% |
✓ |
Kim JA, et al. (2014) [32] |
22 |
22.72% |
63.5 |
Deletion 6q21 |
37% |
x |
t (14q32) |
18% |
x |
Deletion 17p13 (TP53) |
5% |
x |
| |
|
|
Mutation MYD88 (L265P) |
68% |
✓ |
|
|
|
|
Trisomy 5 |
25% |
x |
Mansoor A, et al. [33] |
12 |
|
|
Monosomy 8 |
25% |
x |
Trisomy 3 |
17% |
x |
Deletion 6q |
50% |
x |
Deletion 3q21 |
17% |
x |
Deletion 13q12 |
17% |
x |
Mori N [16] |
25 |
|
|
Mutation MYD88 L265P |
28% |
✓ |
Monge, et al. (2013) [34] |
42 |
|
|
Gain 6p |
16.6% |
x |
|
|
|
|
Deletion 17p13 |
10% |
✓ |
|
30 |
|
|
Mutation MYD88 L265P |
91% |
✓ |
| |
117 |
|
66.4 |
Trisomy 18 |
14.52% |
x |
Nguyen-Khac, et al. [35] |
141 |
|
|
Deletion 6q |
30.49% |
✓ |
145 |
|
|
Deletion 13q14 |
13.10% |
✓ |
139 |
|
|
Trisomy 4 |
7.91% |
✓ |
140 |
|
|
Deletion 17p13 |
7.85% |
✓ |
139 |
|
|
Deletion 11q22 |
7.19% |
x |
140 |
|
|
Trisomy 12 |
4.28% |
x |
Ocio EMN, et al. (2007) [18] |
102 |
|
68 |
Deletion 6q21 |
39% |
✓ |
|
55 |
|
68 |
Deletion 6q |
7.27% |
x |
Petrikkos L, et al. [36] |
36 |
|
65.5 |
Mutation MYD88 L265P |
67% |
✓ |
Poulain S, et al. (2013) [37] |
67 |
37.30% |
|
Mutation MYD88 (L265P) |
79% |
✓ |
Poulain S, et al. (2013) [21] |
31 |
29% |
|
Deletion 6q |
29% |
✓ |
Trisomy 4 |
23% |
✓ |
Gain Xq28 |
16.40% |
x |
Deletion 11q23.2 |
10% |
x |
Deletion 13q14 |
6.45% |
✓ |
Deletion 17q13 |
6.45% |
✓ |
54 |
|
|
Mutation 17q23 |
9.25% |
x |
Mutation 19q13 |
5.55% |
x |
Poulain S, et al. (2016) [20] |
98
53 |
34,3%
|
67 |
CXCR4
CXCR4 |
24.5%
26.4% |
✓
✓ |
Roccaro AM, et al. (2014) [38] |
131 |
|
|
CXCR4 (C1013G) |
28.2% |
✓ |
Schmidt J, et al. [39] |
47
51 |
|
|
CXCR4
MYD88(L265P) |
36.2%
96% |
✓
✓ |
Schop RF, et al. [40] |
37 |
|
|
Deletion 13q14 |
16.20% |
✓ |
|
40 |
|
|
Deletion 17p13.1 |
15% |
x |
Schop RF, et al. [19] |
38 |
|
|
Deletion 6q |
55% |
✓ |
Schop RF, et al. (2002) [41] |
24 |
|
|
Deletion 6q21 |
42-63% |
x |
Terre C, et al. [42] |
39 |
|
|
Trisomy 4 |
8% |
✓ |
Treon SP, et al. [43] |
175 |
|
|
Mutation MYD88 (L265P) |
90.30% |
✓ |
Mutation CXCR4 |
29.10% |
✓ |
Treon SP, et al. [44] |
257 |
|
|
Deletion 6q |
13% |
✓ |
Treon SP, et al. [45] |
30 |
38,7% |
61 |
Mutation CXCR4
Mutation MYD88 (L265P) |
36,7%
96.6% |
✓
✓ |
Treon SP, et al. [46] |
62
63 |
23.8% |
63 |
Mutation CXCR4
Mutation MYD88 (L265P) |
33.8%
88.9% |
✓
✓ |
Varettoni M, et al. [14] |
58 |
|
|
Mutation MYD88 (L265P) |
100% |
x |
Wong KF, et al. [47] |
1 |
100% |
54 |
Trisomy 3 |
100% |
x |
Xu L, et al. (2013) [48] |
104 |
|
|
Mutation MYD88 (L265P) |
93% |
✓ |
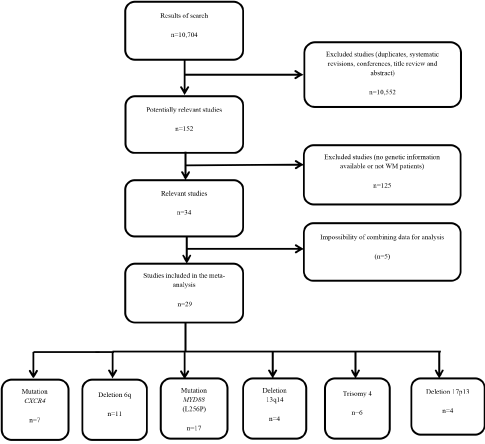
Figure 1. Flow diagram of the literature search
Selected data from the relevant articles subjected to meta-analysis is presented in Table 1. The studies not included in the meta-analysis were those in which there was no possibility to combine data. The meta-analysis consisted of 29 studies and reflected the different genetic variables obtained in each study in addition to the results obtained and the existing relationship between the illness and the different variables. Furthermore, the average age of the patients who participated in the study and their sex was considered, expressed as the percentage of participating women.
Mutation MYD88 (L256P): Of the 29 studies included in the meta-analysis, 17 studied the mutation MYD88 L165P in a total of 1163 patients with WM. The results of the meta-analysis revealed a significant combined incidence rate of 84% (95% confidence intervals (CI): 79, 89; p<0.001), with significant heterogeneity among studies (I2=81.5%, Q=86.6) and no evidence of publication bias (p=0.323) (Figure 2).
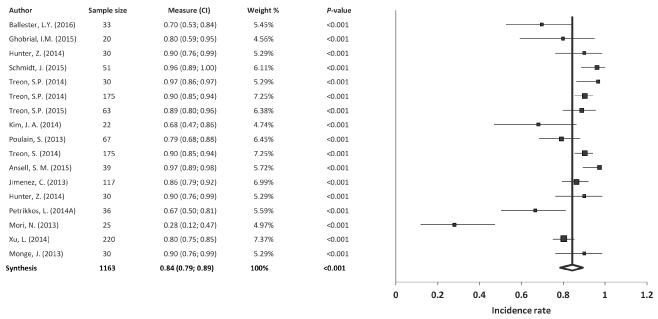
Figure 2. Meta-analysis results of the mutation MYD88
Mutation CXCR4: Of the 29 studies included in the meta-analysis, 7 studied the CXCR4 in 474 patients with WM. The results of the meta-analysis revealed a significant combined incidence rate of 29% (95% confidence intervals (CI): 25, 34; p<0.001), without heterogeneity among studies (I2=0%, Q=3.87) and no evidence of publication bias (p=0.484) (Figure 3).
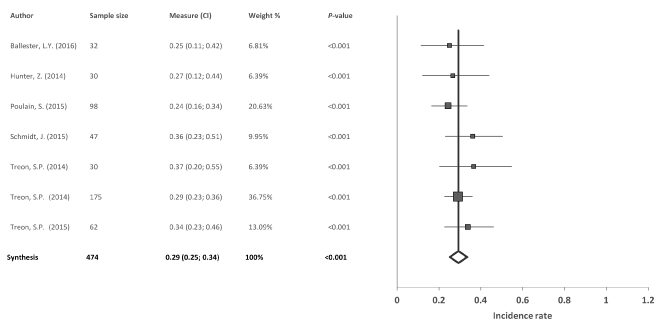
Figure 3. Meta-analysis results of the mutation CXCR4
Deletion 6q: Of the 29 articles used in the meta-analysis, 11 studied the deletion 6q in a total of 836 patients with WM. The results of the meta-analysis revealed a significant combined incidence rate of 36% (95% CI: 26, 45; p<0.001), with high heterogeneity among studies (I2=86.4%, Q=73.4) and evidence of publication bias (p=0.01) (Figure 4).
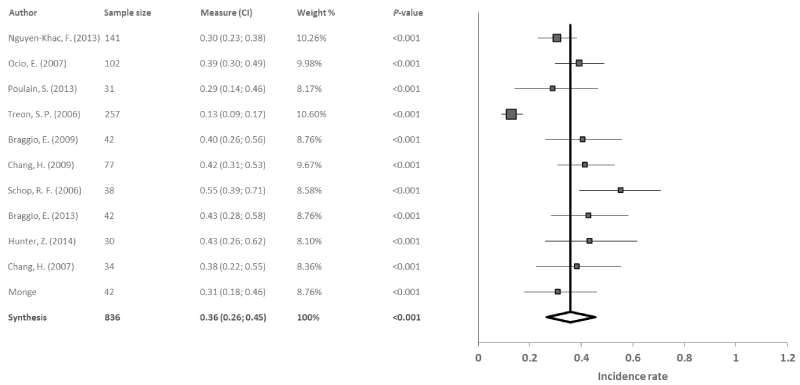
Figure 4. Meta-analysis results of the deletion 6q
Deletion 13q14: Four reports studied deletion 13q14 in 255 patients with WM. The results revealed a significant combined incidence rate of 12% (95% CI: 8, 16; p<0.001), with no evidence of heterogeneity (I2=0%, Q=1.7) or publication bias (p=0.635) among studies (Figure 5).
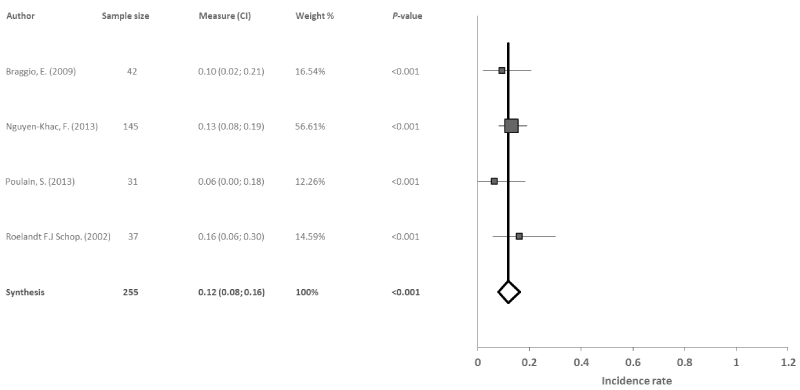
Figure 5. Meta-analysis results of the deletion 13q14
2021 Copyright OAT. All rights reserv
Trisomy 4: Six reports studied trisomy 4 in 323 patients with WM. The results obtained in the meta-analysis revealed a significant combined incidence rate of 14% (95% CI: 8, 20; p<0.001) with heterogeneity among studies (I2=52.4%, Q=10.5) and no evidence of publication bias (p=0.222) (Figure 6).
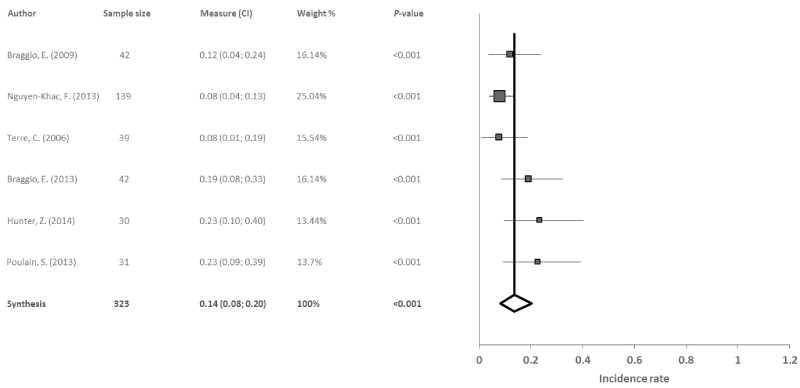
Figure 6. Meta-analysis results of the trisomy 4
Deletion 17p13: Four reports studied deletion 17p13 in 235 patients with WM. The data obtained indicated a significant combined incidence rate of 8% (95% CI: 5, 11; p<0.001), with no evidence of heterogeneity (I2=0%, Q=0.4) or publication bias among studies (p=0.863) (Figure 7).
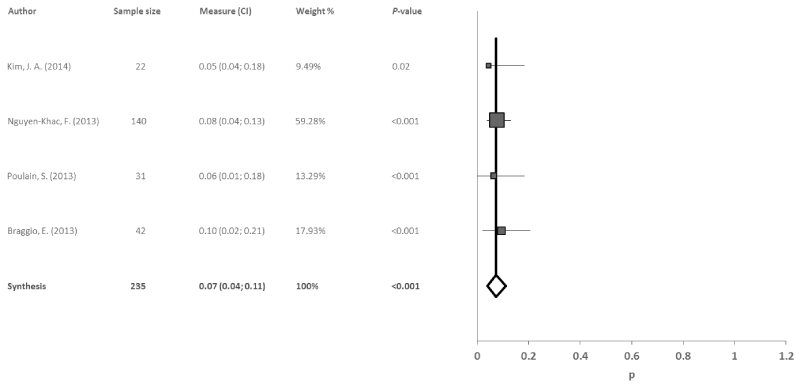
Figure 7. Meta-analysis results of the deletion 17p13
The present meta-analysis shows a high combined incidence rate of 80% (CI 95%: 71, 89) for the MYD88 L256P gene mutation and a combined incidence rate of 34% (CI 95%: 23, 46) for the deletion 6q. The L256P mutation in the MYD88 gene was the most frequent aberration found in patients with WM. MYD88 is an adaptor molecule for the Toll and IL-1R signaling pathways, and is implicated in tumorigenesis through proinflammatory mechanisms [14]. Numerous studies define this variable as the key mutation in WM patients as it is highly prevalent in this population [15]. Excluding the study by Mori, et al. [16], the remaining 11 studies showed an incidence of this mutation that was similar to the combined incidence rate obtained in the meta-analysis. The aforementioned authors reported an incidence rate of 28% as their study had a sensitivity of 10%. The significant heterogeneity observed among the different studies obtained (I2=87.3%, Q=62.9) might reflect the difference in patient numbers in each study and the genotyping methods applied. As the MYD88 mutation is significantly present in the majority of patients with WM, these studies should be extended as they imply a potential advance in the research for possible antigenic therapies to treat this pathology and might also constitute a basis for diagnosing the illness (39).
The studies included in the meta-analysis of deletion 6q presented a combined incidence rate of 34%. Deletion of the long arm of chromosome 6 (6q) is the most frequent cytogenetic abnormality in WM. Chromosome 6 contains the major histocompatibility complex genes, which are responsible for antigen recognition and immune responses [17]. All of the studies analysed in the meta-analysis showed similar incidence rates, although the study by Ocio, et al. [18] used two different techniques to identify abnormalities, conventional cytogenetics and fluorescence in situ hybridization (FISH). While FISH constitutes an efficient technique for the cytogenetic analysis of WM, its use as a prognostic tool is still unknown. Studies such as the one of Schop, et al. [19], showed a high incidence rate of 55%. For this study in particular, to obtain greater accuracy in the criteria of diagnosis, the inclusion criteria included serum IgM ≥1,500 mg·dl-1 and clonal lymphoplasmacytic infiltration comprising at least 20% of the mononuclear cells in the bone marrow biopsy, and therefore the incidence they reported contrasts considerably with the other studies. Heterogeneity was apparent in these studies (I2=90.6%, Q=106.1) given that the criteria for patient diagnosis were different in each study. Furthermore, there was a large variability in sample size between the included cases. Nonetheless, deletion 6q is a significant genetic variable in patients with WM and its study should be of great importance for the development of a new therapeutic target.
Seven studied were included in the meta-analysis assessing the CXCR4 mutation. From 474 patients, the combined incidence rate that patients who presented the mutation was 29%. The CXCR4 mutation has been previously defined as a specific clinic-biological and genomic signature associated to adverse prognosis in WM patients due to its role as a mechanism of resistance to several agents [20].
Four studies were included in the meta-analysis for the deletion 13q14 with a total of 255 patients. No heterogeneity was found between the 4 studies (I2=0%, Q=1.694). The combined incidence rate was significant at 12%, and was similar to the individual studies with the exception of the study by Poulain, et al. [21], which showed an incidence rate of 6% since the technique used for the detection of chromosome alterations was limited regarding the localization of these types of genetic anomalies. This deletion is therefore considered as a potential starting point for identifying new genetic variables for WM, which may lead to new treatment possibilities for patients who present this type of deletion. Also, it offers a basis for future studies to build on, with the aim of identifying other genetic factors that can help in the pathogenesis of the illness.
From the 5 studies of trisomy 4 in WM, the combined incidence rate was also 12%. The values found in the studies tended to be similar, with the exception of the study by Hunter, et al. [22] where the incidence was above average at 23%. Heterogeneity was found in the studies (I2=48.52%, Q=7.770), which could be due to the variability in the number of patients in the different studies and the gender difference. Nevertheless, more research is needed to determine the relative impact of these alterations in WM, as well as the clinical response of these patients to treatment. The trisomy 4 in these patients is identified in persons at high-risk for the illness, and is associated on many occasions with trisomy 18 [19]. Thus, new diagnostic techniques are required for identifying these types of variables, to enable the development of adequate procedures for new treatments.
Finally, the deletion 17p13 showed a combined incidence rate of 8% in the meta-analysis, which was similar to the incidence found in each individual study. Furthermore, no heterogeneity was observed (I2=0%, Q=0.6). The deletion 17p13 is repeatedly present in patients in an advanced stage of the illness and generally presents a low incidence. Patients with this deletion typically have short-term disease-free progression [9]. Although the deletion 17p13 shows a lower incidence rate than other the genetic variables studied, it plays an important role in WM. As it is especially prevalent in advanced stages of the illness, it could be very useful for potential therapies encompassing different treatment regimens.
In summary, different genetic variables play a fundamental role in the pathogenesis of WM. The development and evolution of new diagnostic techniques for the illness and the identification of all the possible genetic factors through DNA sequencing and genome mapping should enable us to establish a basis for new treatment approaches for this illness. The results obtained in the present study show clear evidence of relationships between the different genetic factors and the development of WM, particularly for the MYD88 mutation; however, further extensive investigation is still required on the study of this illness.
The most relevant discovery of this meta-analysis is the identification of the mutation MYD88 (L265P) as a universal marker of the illness. This discovery not only identifies MYD88 as a biomarker to differentiate WM from other malignant B-cell neoplasms, but also as a potential target for the development of more specific therapeutic approaches.
Research by A. Lucia is supported by grants from the Spanish Ministry of Economy and Competitiveness [Fondo de Investigaciones Sanitarias (FIS) and Fondos Feder, grant number [PI15/00558]. The funding source had NO role in: study design, collection, analysis, interpretation of data or writing of the report, and decision to submit the paper for publication.
References
- Hodge LS, Novak AJ, Grote DM, Braggio E, Ketterling RP, et al. (2011) Establishment and characterization of a novel Waldenstrom macroglobulinemia cell line, MWCL-1. Blood 117: e190-197. [Crossref]
- Berger R, Nguyen-Khac F (2008) Chromosomal abnormalities and Waldenstrom macroglobulinemia. Pathol Biol (Paris) 56: 400-406.
- Kristinsson SY, Bjorkholm M, Goldin LR (2008) Risk of lymphoproliferative disorders among first-degree relatives of lymphoplasmacytic lymphoma/Waldenstrom macroglobulinemia patients: a population-based study in Sweden. Blood 112: 3052-3056.
- Treon SP (2009) How I treat Waldenström macroglobulinemia. Blood 114: 2375-2385. [Crossref]
- Merchionne F, Procaccio P, Dammacco F (2011) Waldenstrom's macroglobulinemia. An overview of its clinical, biochemical, immunological and therapeutic features and our series of 121 patients collected in a single center. Crit Rev Oncol Hematol 80: 87-99.
- Braggio E, Keats JJ, Leleu X (2009) Identification of copy number abnormalities and inactivating mutations in two negative regulators of nuclear factor-kappaB signaling pathways in Waldenstrom's macroglobulinemia. Cancer Res 69: 3579-3588.
- Hatjiharissi E, Ngo H, Leontovich AA (2007) Proteomic analysis of waldenstrom macroglobulinemia. Cancer Res 67: 3777-3784.
- Buchsbaum MS, Gillin JC, Wu J (1989) Regional cerebral glucose metabolic rate in human sleep assessed by positron emission tomography. Life Sci 45: 1349-1356.
- Bianchi G, Sacco A, Kumar S (2013) Candidate genes of Waldenstrom's macroglobulinemia: current evidence and research. Appl Clin Genet 6: 33-42.
- Grass S, Preuss KD, Wikowicz A (2011) Hyperphosphorylated paratarg-7: a new molecularly defined risk factor for monoclonal gammopathy of undetermined significance of the IgM type and Waldenstrom macroglobulinemia. Blood 117: 2918-2923.
- Royer RH, Koshiol J, Giambarresi TR (2010) Differential characteristics of Waldenstrom macroglobulinemia according to patterns of familial aggregation. Blood 115: 4464-4471.
- Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg 8: 336-341.
- Nyaga VN, Arbyn M, Aerts M (2014) Metaprop: a Stata command to perform meta-analysis of binomial data. Arch Public Health 72: 39. [Crossref]
- Varettoni M, Arcaini L, Zibellini S (2013) Prevalence and clinical significance of the MYD88 (L265P) somatic mutation in Waldenstrom's macroglobulinemia and related lymphoid neoplasms. Blood 121: 2522-2528.
- Xu L, Hunter ZR, Yang G (2014) Detection of MYD88 L265P in peripheral blood of patients with Waldenstrom's Macroglobulinemia and IgM monoclonal gammopathy of undetermined significance. Leukemia 28: 1698-704.
- Mori N, Ohwashi M, Yoshinaga K, Mitsuhashi K, Tanaka N, et al. (2013) L265P mutation of the MYD88 gene is frequent in Waldenström's macroglobulinemia and its absence in myeloma. PLoS One 8: e80088. [Crossref]
- Trowsdale J, Knight JC (2013) Major histocompatibility complex genomics and human disease. Annual review of genomics and human genetics 14: 301-323.
- Ocio EM, Schop RF, Gonzalez B, Van Wier SA, Hernandez-Rivas JM, et al. (2007) 6q deletion in Waldenström macroglobulinemia is associated with features of adverse prognosis. Br J Haematol 136: 80-86. [Crossref]
- Schop RF, Van Wier SA, Xu R (2006) 6q deletion discriminates Waldenstrom macroglobulinemia from IgM monoclonal gammopathy of undetermined significance. Cancer Genet Cytogenet 169: 150-153.
- Poulain S, Roumier C, Venet-Caillault A (2016) Genomic Landscape of CXCR4 Mutations in Waldenstrom Macroglobulinemia. Clin Cancer Res 22: 1480-1488.
- Poulain S, Roumier C, Galiegue-Zouitina S (2013) Genome wide SNP array identified multiple mechanisms of genetic changes in Waldenstrom macroglobulinemia. Am J Hematol 88: 948-54.
- Hunter ZR, Xu L, Yang G (2014) The genomic landscape of Waldenstrom macroglobulinemia is characterized by highly recurring MYD88 and WHIM-like CXCR4 mutations, and small somatic deletions associated with B-cell lymphomagenesis. Blood 123: 1637-1646.
- Ansell SM, Hodge LS, Secreto FJ (2014) Activation of TAK1 by MYD88 L265P drives malignant B-cell growth in non-Hodgkin lymphoma. Blood 4: 2.
- Rivera AI, Li MM, Beltran G (2002) Trisomy 4 as the sole cytogenetic abnormality in a Waldenström macroglobulinemia. Cancer genetics and cytogenetics 133: 2.
- Ballester LY, Loghavi S, Kanagal-Shamanna R (2016) Clinical Validation of a CXCR4 Mutation Screening Assay for Waldenstrom Macroglobulinemia. Clinical lymphoma, myeloma & leukemia 16: 395-403.
- Braggio E, Keats JJ, Leleu X (2009) High-resolution genomic analysis in Waldenstrom's macroglobulinemia identifies disease-specific and common abnormalities with marginal zone lymphomas. Clinical lymphoma & myeloma 9: 39-42.
- Braggio E, Fonseca R. Genomic abnormalities of Waldenstrom macroglobulinemia and related low-grade B-cell lymphomas. Clinical lymphoma, myeloma & leukemia 13: 198-201.
- Chang H, Qi C, Trieu Y (2009) Prognostic relevance of 6q deletion in Waldenstrom's macroglobulinemia: a multicenter study. Clinical lymphoma & myeloma 9: 36-8.
- Chang H, Qi X, Xu W (2007) Analysis of 6q deletion in Waldenstrom macroglobulinemia. European journal of haematology 79: 244-247.
- Ghobrial IM, Redd R, Armand P (2015) Phase I/II trial of everolimus in combination with bortezomib and rituximab (RVR) in relapsed/refractory Waldenstrom macroglobulinemia. Leukemia 29: 2338-2346.
- Jiménez C, Sebastián E, Chillón MC, Giraldo P, Mariano Hernández J, et al. (2013) MYD88 L265P is a marker highly characteristic of, but not restricted to, Waldenström's macroglobulinemia. Leukemia 27: 1722-1728. [Crossref]
- Kim JA, Im K, Park SN (2014) MYD88 L265P mutations are correlated with 6q deletion in Korean patients with Waldenstrom macroglobulinemia. Biomed Res Int 2014: 363540.
- Mansoor A, Medeiros LJ, Weber DM (2001) Cytogenetic findings in lymphoplasmacytic lymphoma/Waldenstrom macroglobulinemia. Chromosomal abnormalities are associated with the polymorphous subtype and an aggressive clinical course. Am J Clin Pathol 116: 543-549.
- Monge J, Braggio E, Ansell SM (2013) Genetic factors and pathogenesis of Waldenstrom's macroglobulinemia. Current oncology reports 15: 450-456.
- Nguyen-Khac F, Lambert J, Chapiro E (2013) Chromosomal aberrations and their prognostic value in a series of 174 untreated patients with Waldenstrom's macroglobulinemia. Haematologica 98: 649-654.
- Petrikkos L, Kyrtsonis MC, Roumelioti M, Georgiou G, Efthymiou A, et al. (2014) Clonotypic analysis of immunoglobulin heavy chain sequences in patients with Waldenström's macroglobulinemia: Correlation with MYD88 L265P somatic mutation status, clinical features, and outcome. Biomed Res Int 2014:809103.
- Poulain S, Roumier C, Decambron A, Renneville A, Herbaux C, et al. (2013) MYD88 L265P mutation in Waldenstrom macroglobulinemia. Blood 121: 4504-4511. [Crossref]
- Roccaro AM, Sacco A, Jimenez C, Maiso P, Moschetta M, et al. C1013G/CXCR4 acts as a driver mutation of tumor progression and modulator of drug resistance in lymphoplasmacytic lymphoma. Blood 123: 4120-4131.
- Schmidt J, Federmann B, Schindler N, Steinhilber J, Bonzheim I, et al. (2015) MYD88 L265P and CXCR4 mutations in lymphoplasmacytic lymphoma identify cases with high disease activity. Br J Haematol 169: 795-803.
- Schop RFJ, Jalal SM, Wier SAV (2002) Deletions of 17p13.1 and 13q14 are uncommon in Waldenström macroglobulinemia clonal cells and mostly seen at the time of disease progression. Cancer genetics and cytogenetics 132: 5.
- chop RF, Kuehl WM, Van Wier SA, Ahmann GJ, Price-Troska T, et al. (2002) Waldenstrom macroglobulinemia neoplastic cells lack immunoglobulin heavy chain locus translocations but have frequent 6q deletions. Blood 100: 2996-3001.
- Terre C, Nguyen-Khac F, Barin C, et al. (2006) Trisomy 4, a new chromosomal abnormality in Waldenstrom's macroglobulinemia: a study of 39 cases. Leukemia 20: 1634-1636.
- Treon SP, Cao Y, Xu L, Yang G, Liu X, et al. (2014) Somatic mutations in MYD88 and CXCR4 are determinants of clinical presentation and overall survival in Waldenstrom macroglobulinemia. Blood 123: 2791-2796.
- Treon SP, Hunter ZR, Aggarwal A, Ewen EP, Masota S, et al. (2006) Characterization of familial Waldenstrom's macroglobulinemia. Ann Oncol 17: 488-494. [Crossref]
- Zreon SP, Tripsas CK, Meid K, et al. Carfilzomib, rituximab, and dexamethasone (CaRD) treatment offers a neuropathy-sparing approach for treating Waldenstrom's macroglobulinemia. Blood 124: 503-510.
- 46. Treon SP, Tripsas CK, Meid K (2015) Ibrutinib in previously treated Waldenstrom's macroglobulinemia. N Engl J Med 372: 1430-1440.
- Wong KF, Kwong YL, Wong TK (1995) Concomitant partial tetrasomy 3q and trisomy 18 in waldenstrom macroglobulinemia. Cancer genetics and cytogenetics 81: 2.
- 48. Xu L, Hunter ZR, Yang G, et al. MYD88 L265P in Waldenstrom macroglobulinemia, immunoglobulin M monoclonal gammopathy, and other B-cell lymphoproliferative disorders using conventional and quantitative allele-specific polymerase chain reaction. Blood. 2013 Mar 14;121(11):2051-8. doi: 10.1182/blood-2012-09-454355. PubMed PMID: 23321251; PubMed Central PMCID: PMC3596964.







