Forkhead box F2 (FOXF2) functions as a transcription factor and is critically involved in programming organogenesis and regulating epithelial-to-mesenchymal transition (EMT) and cell proliferation. We recently have revealed that FOXF2 can exert distinct functional effects on different molecular subtypes of breast cancer. We found that FOXF2 expression is epigenetically silenced in luminal breast cancers due to its tumor-suppressive role in DNA replication regulation. In contrast, FOXF2 is overexpressed in basal-like triple-negative breast cancers (TNBCs) due to its oncogenic role in promoting EMT. Although our and other studies have shown that FOXF2 dysregulation is critical for tumorigenesis of various tissue types, the role of FOXF2 in metabolic rewiring of cancer remains unknown. In this study, we analyzed our previous microarray data to understand the metabolic role of FOXF2 in non-cancerous and cancerous breast epithelial cells. Our studies showed that in non-cancerous breast epithelial cells FOXF2 can also play a dual role either in tumor suppression or in tumor promotion through regulating expression of tumor-suppressive and oncogenic metabolic genes. Furthermore, we found that FOXF2-regulated metabolic genes are not conserved between non-cancerous and cancerous breast epithelial cells and FOXF2 is involved in metabolic rewiring in breast cancer cells. This is the first report to explore the metabolic function of FOXF2 in breast cancer.
FOXF2, metabolism, breast cancer
Metabolic reprogramming has been recognized as a hallmark of cancer due to its critical role in tumorigenesis [1]. For breast cancer, a large number of studies have shown that breast cancer cells rewire their various metabolisms to fulfill the demands of survival, proliferation, invasion and metastasis. Given that breast cancer is a heterogeneous disease, metabolic rewiring is differentially exhibited among different breast cancers or even within a breast tumor that has been known to have intra-tumor heterogeneity. Clinically, breast cancers have been commonly classified according to expression of three receptors: estrogen receptor (ER), progesterone receptor (PR), and HER2. This classification system has guided targeted treatment regimens, such as endocrine therapy for ER-positive breast cancer and HER2-targeting therapy for HER2-postiive breast cancer for decades. Due to the development of microarray technology, genome-wide gene expression profiling has been exploited to molecularly classify heterogeneous breast cancers into at least five subtypes, including normal breast-like, luminal A (ER+ and/or PR ± HER2– with a low Ki67 index), luminal B (ER+ and/or PR ± HER2+ or HER2– with a high Ki67 index), HER2-positive (ER–/PR–/HER2+) and basal-like/triple negative (ER–/PR–/HER2–) [2,3]. Among these molecular subtypes, basal-like triple-negative breast cancer (TNBC), characterized by its lack of ER, PR and HER2, is aggressive and lacks targeted therapies [3]. Basal-like TNBCs show poor clinical outcomes due to its high tumor grade, increased rate of proliferation and metastasis, and frequent recurrence [3]. For exploring the therapeutic potential of metabolic targeting, numerous studies were conducted to examine how cellular metabolisms are rewired in different molecular subtypes of breast cancer. Their findings have shown that each subtype of breast cancer displays distinct metabolic alterations [4]. For example, TNBCs and HER2-positive breast cancers manifest higher glycolytic activity (Warburg effect) and glutamine metabolism compared to luminal breast cancers. Understanding of metabolic heterogeneity would help guide breast cancer therapy and improve patient outcomes. Despite the importance of metabolic reprogramming in breast cancer development, what molecular regulators are engaged in this metabolic rewiring in breast cancer remains largely unknown.
The forkhead box (FOX) family genes encode evolutionally conserved transcription factors functionally involved in modulating global gene expression to regulate various biological processes, such as embryogenesis, pattern formation, immune responses and aging in multicellular organisms, and cell cycle progression, DNA damage responses and cellular metabolism in cells. FOX proteins bind genomic DNA as a monomer through their forkhead domain to transcriptionally regulate their target genes. Forkhead box F2 (FOXF2), a FOXF subfamily gene member in the FOX gene family, is a key regulator implicated in promoting mesenchymal programming. Embryonic studies have shown that FOXF2 is specifically expressed in the mesenchyme. These FOXF2-positive mesenchymal tissues are directly adjacent to the ectoderm-derived epithelium that develops into tongue and to the endoderm-derived epithelium that develops into the gastrointestinal (GI) tract, lungs, and genitalia [5]. Animal studies have shown that Foxf2 is essential for organogenesis of palate, tongue and gut as Foxf2-knockout mice have developmental defects in these organs [6-9]. Moreover, studies of Foxf2-deficient mice have revealed that the critical role of Foxf2 in organogenesis involves its function in mediating hedgehog signaling to activate Tgfβ signaling and inhibiting Wnt as well as Fgf18 signaling [7-9]. These Foxf2-regulated signaling pathways are critical for maintaining extracellular matrix (ECM) content and the functions of mesenchyme as well as epithelium for organogenesis [7-9].
Increasing evidence from recent studies has linked FOXF2 dysregulation to a variety of cancers, such as breast, colon, esophageal, lung, liver and prostate cancers [10-19]. Studies of these cancer types have revealed that FOXF2 is aberrantly downregulated in cancer cells through epigenetic silencing mechanisms including DNA methylation of the FOXF2 promoter and targeting by oncogenic microRNAs (e.g. miR-301, miR-182 and miR-519a). Given that restoring FOXF2 expression in FOXF2-deficient cancer cells inhibits cancer cell growth and other tumorigenic features, these studies have indicated that FOXF2 functions as a tumor suppressor. Nevertheless, it has been reported that FOXF2 acts as an oncogenic factor in alveolar rhabdomyosarcoma and lung cancer [14,20]. These controversial findings suggest that FOXF2 can function as either a tumor suppressor gene or an oncogene depending on tissue-context and stage-specific scenarios. Consistent with this proposed view, our recent studies have revealed that FOXF2 has a dual role in breast cancer [19]. We found that FOXF2 acts as a tumor suppressor and is epigenetically silenced in luminal breast cancers, whereas FOXF2 is overexpressed in basal-like TNBCs and functions as an oncogene in this breast cancer subtype [19].
In this study, we focused on the regulatory role of FOXF2 in metabolic gene expression in non-cancerous breast epithelial cells and in basal-like TNBC cells. We analyzed our previous global gene expression microarray datasets [19] and found that rewiring expression of metabolic genes occurred in basal-like TNBC cells when compared to non-cancerous breast epithelial cells and FOXF2 participateds in this metabolic rewiring.
Cell lines and tissue samples
We obtained immortalized, nontumorigenic human mammary epithelial cells (MCF10A) and the basal-like breast cancer cells (MDA-MB-231) from ATCC (American Type Culture Collection, Manassas, VA, USA) and cultured them according to the ATCC online instructions. The molecular subtype classification of breast cancer cell lines used in the study was based on the information of two publications [21,22].
siRNA transfection
siRNA transfections were performed with 20 nM of each siRNA using OligofectamineTM RNAiMAX (Life Technologies Inc.) according to the instructions of the manufacturer. The siRNA sequence for targeting FOXF2 is: 5’-CAACUUCAAUGGGAUUUCU-3’. The FOXF2 siRNA and non-targeting control siRNA (siControl: Catalog No. D-001810-10) were purchased from Dharmacon (Lafayette, CO, USA).
Gene expression profiling
Microarray analysis was performed as previously described [19]. In brief, 48 hours after siRNA transfections, siRNA-transfected MCF10A (or MDA-MB-231) cells were lysed in TRIzol (Life Technologies Inc.) and total RNA was purified. Approximately 20 mg of RNA was treated with RNase-free DNaseI (#2238, Ambion, Ambion Inc., Austin, TX, USA) and subsequently purified using the RNeasy mini kit (#74104, Qiagen, Hilden, Germany). The integrity of purified DNaseI-treated RNA was evaluated using the Agilent 2100 Bioanalyzer and the RNA nano 6000 kit (Agilent Technologies, Wilmington, DE, USA). For each sample, Biotinylated cRNA was prepared using the Ambion MessageAmplification kit for Illumina arrays (Ambion Inc.) with an input of 500 ng total RNA. Per sample, 750 ng of the biotinylated cRNA was hybridized onto the Illumina HumanHT-12 v4 Expression BeadChip (Illumina, Inc., San Diego, CA, USA). Hybridized arrays were scanned on an Illumina HiScan microarray scanner. Illumina GenomeStudio was used to transform bead-level data to probe-level intensity values and statistics, which were exported raw data (unfiltered and non-normalized) for bioinformatic analysis. The expression data were quantile normalized using IlluminaGUI in R and log2-transformed, and a rank product analysis was performed using a p-value < 0.05 to identify significant changes of gene expression. The datasets have been deposited in the GEO data repository (http://www.ncbi.nlm.nih.gov/geo, accession number GSE55675).
In silico analysis of gene expression
The Oncomine’s Cancer Microarray Database (http://www.oncomine.org) [23] was used to perform in silico expression analysis of metabolic genes in normal and cancerous breast tissues.
Statistical analysis
The Student’s t-test was used to analyze the significance of difference between two groups of data using the GraphPad Prism software (version 6.0; GraphPad Software, Inc, La Jolla, CA, USA). p < 0.05 was regarded as statistically significant.
FOXF2-regulated metabolic genes in non-cancerous breast epithelial cells
We previously showed that FOXF2 was expressed in normal breast epithelial cells and required for their cell cycle progression and mobility [19]. Compared to normal breast epithelial cells, over 60% of basal-like TNBC cell lines overexpressed FOXF2 [19]. FOXF2 knockdown studies demonstrated that FOXF2 is crucial for anchorage-independent growth, migration and invasion of basal-like TNBC cells [19]. These findings indicate that FOXF2 plays physiological and pathological roles in normal breast epithelial and basal-like TNBC cells, respectively. To reveal the biological roles of FOXF2 in normal breast epithelial and metastatic basal-like TNBC cells, we previously performed global gene expression profiling analyses of FOXF2-knockdown MCF10A (a noncancerous breast epithelial cell line) and MDA-MB-231 (a metastatic basal-like TNBC cell line) cells compared with their respective control siRNA-transfected cells using microarrays [19]. Our microarray-based gene expression profiling studies identified 199 and 309 differentially expressed genes (≥ 2fold) in FOXF2-knockdown MCF10A and MDA-MB-231 cells, respectively [19]. To identify FOXF2-regulated metabolic genes in MCF10A cells, we performed gene ontology enrichment analysis using EnrichNet (http://www.enrichnet.org). Nine metabolic genes were identified to be regulated by FOXF2 (Table 1). Aldehyde dehydrogenase 1 family member A3 (ALDH1A3) is the sole gene that was downregulated in FOXF2-knockdown MCF10A cells, indicating that FOXF2 positively regulates ALDH1A3 expression and negatively regulates expression of remaining 8 metabolic genes in MCF10A cells. The metabolic roles of these genes are summarized in Figure 1.
Table 1. The list of differentially expressed genes identified in FOXF2-knockdown MCF10A cells.
Gene symbol |
Full Gene Name |
MCF10A |
MDA-MB-231 |
Control siRNAa |
FOXF2 siRNAa |
Foldb |
Control siRNAa |
FOXF2 siRNAa |
Foldb |
AKR1C3 |
aldo-keto reductase family 1 member C3 |
2469 |
7372 |
2.99 |
1306 |
1755 |
1.34 |
ALDH1A3 |
aldehyde dehydrogenase 1 family member A3 |
440 |
155 |
0.35 |
55 |
34 |
0.62 |
CYP1B1 |
cytochrome P450 family 1 subfamily B member 1 |
167 |
427 |
2.56 |
839 |
1413 |
1.68 |
HMGCL |
3-hydroxy-3-methylglutaryl-CoA lyase |
62 |
137 |
2.21 |
65 |
70 |
1.08 |
MAOA |
monoamine oxidase A |
508 |
1185 |
2.33 |
225 |
241 |
1.07 |
PANK4 |
pantothenate kinase 4 |
77 |
163 |
2.12 |
106 |
123 |
1.16 |
PRODH |
proline dehydrogenase 1 |
51 |
117 |
2.29 |
ns |
ns |
|
SGPP2 |
sphingosine-1-phosphate phosphatase 2 |
206 |
473 |
2.30 |
33 |
56 |
1.70 |
TPH1 |
tryptophan hydroxylase 1 |
31 |
69 |
2.23 |
80 |
55 |
0.69 |
a Non-specific background values from microarray analysis have been subtracted from expression values. Expression values have been normalized to their housekeeping genes.
b The expression fold is FOXF2 siRNA relative to control siRNA.
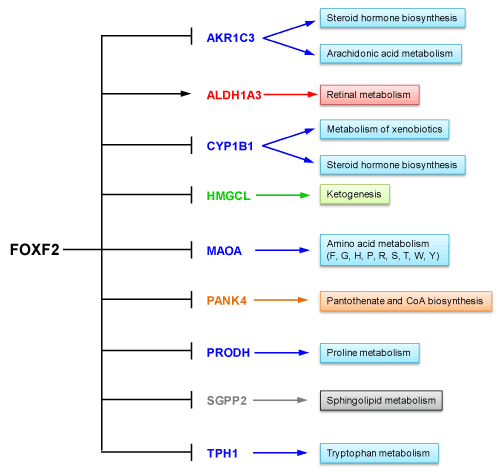
Figure 1. FOXF2 regulates expression of 9 metabolic genes in MCF10A cells. Metabolisms associated with these FOXF2-regulated genes are indicated.
ALDH1A3 is known to preferentially oxidize retinal (retinaldehyde, one form of vitamin A) to retinoic acid (RA). Therefore, ALDH1A3 is one of key enzymes contributing to RA biosynthesis. ALDH1A3 has been reported to be required for ALDH activity, an important indicator for detecting cancer stem cells (CSCs) using the Aldefluor assay, and RA biosynthesis in breast cancer cells [24]. Moreover, ALDH1A3 expression has been correlated significantly with higher grade tumors, proximal metastasis, and higher cancer stage of breast cancers [24]. A study that analyzed breast patient tumors revealed that high levels of ALDH1A3 correlated with expression of RA-inducible genes with retinoic acid response elements (RAREs), poorer patient survival and TNBCs [25]. Our in silico analysis of ALDH1A3 expression in different molecular breast cancer subtypes using Esserman breast cancer datasets retrieved from the Oncomine database (www.oncomine.org) [23] also showed that ALDH1A3 expression levels were higher in basal-like TNBCs than in luminal breast cancers (Figure 2). This result and aforementioned findings from other studies [24,25] suggest that the FOXF2-ALDH1A3 axis may contribute to CSC features, invasiveness and metastasis of FOXF2-overexpressing TNBCs. Interestingly, this FOXF2-ALDH1A3 axis identified from the study of MCF10A cells was lost in MDA-MB-231 cells as this metastatic TNBC line expressed the very low levels of ALDH1A3 (Table 1), consistent with the finding reported elsewhere [24]. This contradictory phenotype may be cell line-specific. Nevertheless, ectopic ALDH1A3 overexpression or RA treatment has been reported to promote xenograft tumor growth and metastasis of MDA-MB-231 cells [25].
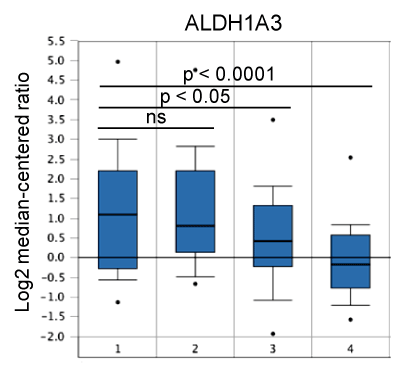
Figure 2. Expression of ALDH1A3 in different molecular subtypes of breast cancer. 1: Basal-like (n=43); 2: HER2+ (n=21); 3: Luminal A (n=32); 4: Luminal B (n=25).
As shown in Figure 1, FOXF2 may negatively control amino acid metabolism, sphingolipid metabolism, steroid hormone biosynthesis, arachidonic acid metabolism, coenzyme A (CoA) biosynthesis and ketogenesis through its negatively regulatory effect on expression of 8 metabolic genes, including aldo-keto reductase family 1 member C3 (AKR1C3), cytochrome P450 family 1 subfamily B member 1 (CYP1B1), 3-hydroxy-3-methylglutaryl-CoA lyase (HMGCL), monoamine oxidase A (MAOA), pantothenate kinase 4 (PANK4), proline dehydrogenase 1 (PRODH), sphingosine-1-phosphate phosphatase 2 (SGPP2), and tryptophan hydroxylase 1 (TPH1). Among these genes, four genes (AKR1C3, MAOA, PRODH, SGPP2) were expressed at a lower level in MDA-MB-231 cells than in MCF10A cells and their expression was not significantly regulated by FOXF2 in MDA-MB-231 cells (Table 1). These results suggest that the functions of these four genes may have a negative impact on the development of TNBC and some alternative pathological mechanisms develop for replacing FOXF2 to further downregulate expression of these four genes. In line with this viewpoint, the in silico expression analysis of these four genes in breast cancer using TCGA and Gluck breast cancer datasets retrieved from the Oncomine database showed expression of AKR1C3 and MAOA was generally downregulated in invasive breast carcinomas, and PRODH was expressed at a lower level in basal-like TNBCs than in luminal breast cancer subtypes (Figure 3). Indeed, AKR1C3 (also known as 17beta-hydroxysteroid dehydrogenase type 5 or 3alpha-hydroxysteroid dehydrogenase type 2), which functions as a 3-keto, 17-keto and 20-ketosteroid reductase and as a 3alpha-, 17beta- and 20alpha-hydroxysteroid oxidase, has been reported to be downregulated in breast cancer due to its role in producing steroid metabolites with the inhibitory effect on mitogenesis and metastasis of breast cancer cells [26]. Consistent with our in silico analysis data (Figure 3), MAOA, a mitochondrial enzyme involved in catalyzing the oxidative deamination of amines, has been reported to be under-expressed in multiple cancer types (including breast cancer) compared to their corresponding normal tissues [27]. Frequent downregulation of MAOA in cancer may be related to its role in amino acid metabolism, which depletes amino acid resources for protein synthesis and limits cell growth of cancer cells that highly demand protein synthesis for their proliferation. PRODH is a flavin-dependent enzyme catalyzing the conversion of proline into Δ(1) -pyrroline-5-carboxylate (P5C). This PRODH-mediated enzymatic reaction produces reactive oxygen species (ROS), which induces intrinsic and extrinsic apoptotic pathways [28]. In addition, PRODH, a p53 target gene, is involved in negatively regulating multiple oncogenic factors and pathways such as HIF-1α transcriptional activity, the MAPK pathway, cyclooxygenase-2, epidermal growth factor receptor and Wnt/β-catenin signaling [28]. Therefore, PRODH downregulation provides advantages to breast tumorigenesis. Sphingolipids are one of cell membrane components and downregulation of sphingolipid-metabolizing enzyme SGPP2 in TNBC cells may help the demand of cell membrane building during cell proliferation. These multiple lines of evidence suggest that negative regulation of these four genes by FOXF2 may provide initial advantages to TNBC development before alternative mechanisms develop to further inhibit their expression. These findings suggest that FOXF2 may play an oncogenic role in early development of basal-like TNBC by negatively regulating these tumor-suppressive metabolic genes.
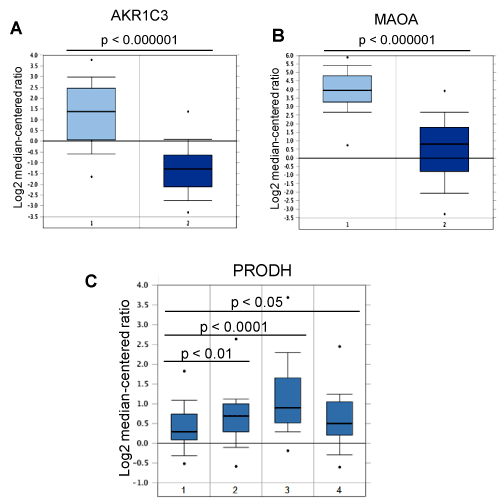
Figure 3. Expression of AKR1C3, MAOA and PRODH in breast cancer. (A) AKR1C3 expression is downregulated in breast cancer. 1: Normal breast (n=61); 2: Invasive breast carcinoma (n=76). (B) MAOA expression is downregulated in breast cancer. 1: Normal breast (n=61); 2: Invasive breast carcinoma (n=76). (C) Expression of PRODH in different molecular subtypes of breast cancer. 1: Basal-like (n=45); 2: HER2+ (n=21); 3: Luminal A (n=46); 4: Luminal B (n=25).
Another potential oncogenic function of FOXF2 is its role in negative regulation of HMGCL expression in MCF10A cells (Table 1). HMGCL, an essential enzyme in ketogenesis, has been shown to be downregulated in nasopharyngeal carcinoma (NPC) [29]. Ectopic expression of HMGCL restored β-hydroxybutyrate (β-HB, a ketone metabolite) levels, inhibited proliferation and colony formation of NPC cells in vitro and suppressed NPC tumorigenicity in vivo [29]. HMGCL also impaired the migration and invasion of NPC cells in vitro through activation of mesenchymal-epithelial transition (MET) [29]. Moreover, extracellular β-HB supply mimicked HMGCL's effect to attenuate the proliferation and migration of NPC cells [29]. The suppressive role of both intra- and extracellular β-HB in NPC relied on reactive oxygen species (ROS) generation [29]. These findings suggest negative regulation of HMGCL expression by FOXF2 may generate an advantageous scenario for basal-like TNBC development. Although MDA-MB-231 cells expressed the same level of HMGCL as that observed in MCF10A cells, HMGCL expression in MDA-MB-231 cells became FOXF2-independent (Table 1), suggesting that some TNBCs develop alternative mechanisms for replacing FOXF2 to maintain HMGCL expression at an adequate level.
Three FOXF2-regulated metabolic genes (CYP1B1, PANK4, TPH1) identified in the microarray study of MCF10A cells were expressed at a higher level in MDA-MB-231 cells when compared to MCF10A cells and their expression became more FOXF2-independent in MDA-MB-231 cells (Table 1). These results suggest that these three metabolic genes may have oncogenic roles in basal-like TNBC development. In line with our finding, CYP1B1, a heme-thiolate monooxygenase mainly expressed in endocrine-regulated tissues like breast, has been shown to be overexpressed in breast cancer, including basal-like TNBCs [30]. CYP1B1 has an oncogenic role in cancer due to its role in the metabolism of 17β-estradiol (E2) and E2-like molecules, which causes DNA adducts and generates free radicals leading to DNA damage and tumorigenesis in different tissues like breast [30]. Consistently, TPH1, an enzyme involved in serotonin (5-hydroxytryptamine or 5-HT) biosynthesis via catalyzing tryptophan metabolism, has been shown to be overexpressed in basal-like TNBC cell lines like MDA-MB-231 [31]. In line with this finding, 5-HT treatment promoted invasion and proliferation of MDA-MB-231 cells via 5-HT7 receptor [31]. These findings together imply that FOXF2 may also have a tumor-suppressive role in TNBC development via its negative regulation of CYP1B1 and TPH1.
FOXF2-regulated metabolic genes in basal-like breast cancer cells
Through gene ontology enrichment analysis, we identified 20 upregulated metabolic genes in FOXF2-knockdown MDA-MB-231 cells (Table 2). The metabolic roles of these 20 genes are summarized in Figure 4. In contrast to expression data from studies of MDA-MB-231 cells, expression of the majority of these genes was FOXF2-independent in MCF10A cells. These findings indicated that FOXF2-dependent gene expression regulation was significantly altered in basal-like TNBC cells. Interestingly, after we compared MDA-MB-231 microarray datasets of these 20 genes with their respective MCF10A datasets, we found that these genes can be classified into two different groups based on their expression patterns in both cell lines. The first gene group encompassing 10 genes (including ADPRM, CA9, G6PD, HMOX1, LYPLA2, MTHFR, NDST1, PIGK, PIP5K1C, UST) displayed aberrant upregulation in MDA-MB-231 cells when FOXF2 was knocked down by siRNA but showed similar expression levels in both control siRNA-transfected MDA-MB-231 and MCF10A cells (Table 2). This observation suggests that MDA-MB-231 cells need FOXF2 to negatively regulate expression of these 10 genes for maintaining adequate activities of these 10-gene-involved cellular metabolisms. This finding prompted us to hypothesize that during basal-like TNBC development, cancer cells require FOXF2 to suppress aberrant activation of some metabolisms for preventing their harmful effects.
Table 2. The list of differentially expressed genes identified in FOXF2-knockdown MDA-MB-231 cells.
Gene symbol |
Full Gene Name |
MDA-MB-231 |
MCF10A |
Control siRNAa |
FOXF2 siRNAa |
Foldb |
Control siRNAa |
FOXF2 siRNAa |
Foldb |
ALDH1B1 |
aldehyde dehydrogenase 1 family member B1 |
126 |
273 |
2.16 |
247 |
276 |
1.12 |
ADPRM |
ADP-ribose/CDP-alcohol diphosphatase, manganese dependent |
64 |
152 |
2.38 |
86 |
114 |
1.33 |
CA9 |
carbonic anhydrase 9 |
69 |
230 |
3.33 |
54 |
57 |
1.06 |
CAD |
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
179 |
401 |
2.24 |
413 |
411 |
1.00 |
G6PD |
glucose-6-phosphate dehydrogenase |
773 |
1686 |
2.18 |
649 |
793 |
1.22 |
GALNTL4 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 4 |
122 |
271 |
2.22 |
524 |
452 |
0.86 |
GDA |
guanine deaminase |
35 |
78 |
2.23 |
78 |
86 |
1.10 |
HMOX1 |
heme oxygenase 1 |
67 |
465 |
6.94 |
44 |
83 |
1.89 |
LYPLA2 |
lysophospholipase II |
333 |
758 |
2.28 |
460 |
461 |
1.00 |
MTHFR |
methylenetetrahydrofolate reductase |
44 |
111 |
2.52 |
66 |
89 |
1.35 |
NDST1 |
N-deacetylase and N-sulfotransferase 1 |
197 |
437 |
2.22 |
250 |
278 |
1.11 |
NDUFS2 |
NADH:ubiquinone oxidoreductase core subunit S2 |
73 |
155 |
2.12 |
192 |
224 |
1.17 |
PIGK |
phosphatidylinositol glycan anchor biosynthesis class K |
254 |
520 |
2.05 |
190 |
318 |
1.67 |
PIK3C2A |
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 alpha |
60 |
121 |
2.02 |
109 |
75 |
0.69 |
PIK3C2B |
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
20 |
63 |
3.15 |
80 |
83 |
1.04 |
PIP5K1C |
phosphatidylinositol-4-phosphate 5-kinase type 1 gamma |
216 |
506 |
2.34 |
266 |
398 |
1.50 |
PLA2G12A |
phospholipase A2 group XIIA |
24 |
66 |
2.75 |
71 |
69 |
0.97 |
PLD2 |
phospholipase D2 |
34 |
106 |
3.12 |
172 |
201 |
1.17 |
PMVK |
phosphomevalonate kinase |
62 |
152 |
2.45 |
249 |
303 |
1.22 |
UST |
uronyl 2-sulfotransferase |
60 |
144 |
2.40 |
98 |
129 |
1.32 |
a Non-specific background values from microarray analysis have been subtracted from expression values. Expression values have been normalized to their housekeeping genes.
b The expression fold is FOXF2 siRNA relative to control siRNA.
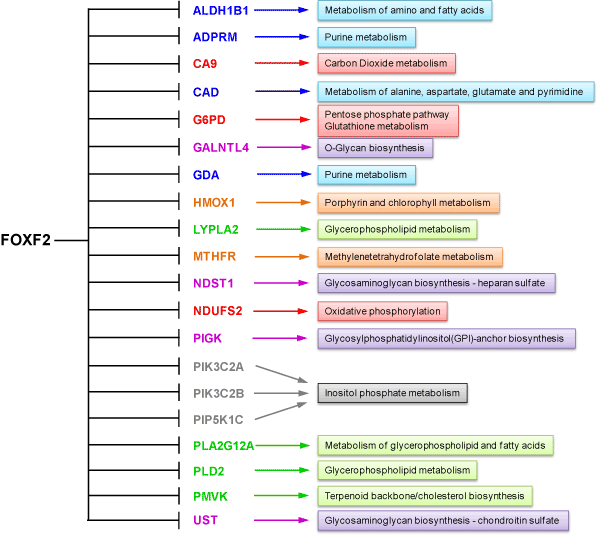
Figure 4. FOXF2 regulates expression of 20 metabolic genes in MDA-MB-231 cells. Metabolisms associated with these FOXF2-regulated genes are indicated.
The second gene group encompassing 10 genes (including ALDH1B1, CAD, GALNTL4, GDA, NDUFS2, PIK3C2A, PIK3C2B, PLA2G12A, PLD2, PMVK) manifested FOXF2-dependent under-expression in MDA-MB-231 cells when compared to MCF10A datasets (Table 2). This observation suggests that high expression levels of this gene group are unfavorable and thus their expression is preferentially downregulated by FOXF2 in MDA-MB-231 cells. To reveal whether this observation is relevant to basal-like TNBCs, we performed in silico expression analysis of these 10 genes using Esserman and Gluck breast cancer datasets retrieved from the Oncomine database. The analysis results showed that PLA2G12A and PMVK were expressed at a lower level in basal-like TNBCs compared to luminal breast cancers (Figure 5). These findings suggest that these two metabolic genes may play tumor-suppressive roles or have unfavorable effects in basal-like TNBC development. PLA2G12A belongs to the group XII of secreted phospholipases A2 (sPLA2) enzymes that function to liberate arachidonic acid from phospholipids for production of eicosanoids and exert a variety of physiologic and pathologic effects. The functional role of PLA2G12A in breast cancer remains elusive and needs further investigation. PMVK is a metabolic enzyme involved in the mevalonate pathway, which is required for the generation of several fundamental end-products including cholesterol and isoprenoids. Protein prenyltransferases exploit the isoprenoid metabolites such as farnesyl diphosphate (FPP) and geranylgeranyl diphosphate (GGPP) to catalyze post-translational isoprenylation of cysteine residues at the C termini of a wide variety of proteins, which is an important mechanism to modulate protein functionality. The mevalonate pathway has been shown to be activated in cancer and ectopic expression of a rate-limiting enzyme called hydroxymethylglutaryl coenzyme A reductase (HMGCR) in this pathway led to transformation of mouse embryonic fibroblasts [32]. It will be interesting in the future to reveal why PMVK is preferentially downregulated in basal-like TNBCs and whether its downregulation affects the mevalonate pathway in basal-like TNBCs. Although the role of the mevalonate pathway in basal-like TNBC is unclear, a related study showed that MDA-MB-231 cells efficiently used exogenous isoprenols for protein isoprenylation in independent of the mevalonate pathway [33]. This clue suggests that TNBC cells may be capable of using exogenous isoprenols for their survival and growth even though endogenous isoprenoid biosynthesis is limited.
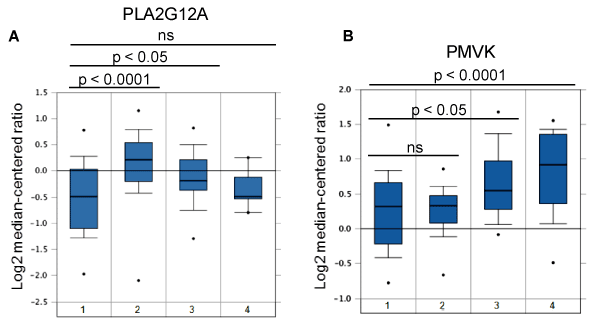
Figure 5. Expression of PLA2G12A and PMVK in breast cancer. (A) Expression of PLA2G12A in different molecular subtypes of breast cancer. 1: Basal-like (n=43); 2: HER2+ (n=21); 3: Luminal A (n=32); 4: Luminal B (n=25). (B) Expression of PMVK in different molecular subtypes of breast cancer. 1: Basal-like (n=45); 2: HER2+ (n=21); 3: Luminal A (n=46); 4: Luminal B (n=25).
Our comprehensive analysis of microarray expression data has built an initial picture for the metabolic role of FOXF2 in breast cancer. We observed that non-cancerous breast epithelial and basal-like TNBC cells displayed distinct FOXF2-regulated metabolic gene expression signatures, indicating that FOXF2 has different metabolic roles in non-cancerous and cancerous breast epithelial cells. This difference likely results from the dramatically altered transcriptional environment in breast cancer cells caused by significant alterations in their genome, epigenome and transcriptome. Under the different transcriptional environment, FOXF2 can display distinct gene target specificity through its association with various transcriptional cofactors. Interestingly, we for the first time revealed that FOXF2 can regulate expression of both tumor-suppressive and oncogenic metabolic genes, in line with our previous discovery of FOXF2's dual function in breast cancer [19]. Due to the critical role of metabolic rewiring in cancer development, identification of molecular regulators involved in this event is crucial for understanding of cancer metabolism and developing novel therapeutic medicine targeting metabolic vulnerabilities in cancer cells. Importantly, our microarray data analysis has unraveled that FOXF2 is a novel molecular regulator critically engaged in metabolic rewiring in basal-like TNBC cells. Moreover, we surprisingly found that FOXF2 mainly inhibits expression of metabolic genes in both non-cancerous breast epithelial and basal-like TNBC cells. Although it is unclear whether FOXF2 regulates these metabolic genes in a direct or indirect manner, this discovery has prompted us to consider that FOXF2-mediated transcription repression may play a critical role in normal and cancer cell metabolisms. Future investigations are needed to decipher the roles of these novel FOXF2-mediated metabolic functions in proliferation, invasiveness and metastasis of basal-like TNBC.
The FOXF2 studies were supported by the National Institutes of Health (grant number: SPORE-1 P50 CA088843-08) and the Department of Defense (grant number: Center of Excellence-W81XWH-04-1-0595) to Dr. Saraswati Sukumar at the Johns Hopkins University.
The author declares no conflict of interest.
- Pavlova NN, Thompson CB (2016) The Emerging Hallmarks of Cancer Metabolism. Cell Metab 23: 27-47. [Crossref]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. (2000) Molecular portraits of human breast tumours. Nature 406: 747-752. [Crossref]
- Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, et al. (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 98: 10869-10874. [Crossref]
- Ogrodzinski MP, Bernard JJ, Lunt SY (2017) Deciphering metabolic rewiring in breast cancer subtypes. Transl Res 189: 105-122. [Crossref]
- Aitola M, Carlsson P, Mahlapuu M, Enerbäck S, Pelto-Huikko M (2000) Forkhead transcription factor FoxF2 is expressed in mesodermal tissues involved in epithelio-mesenchymal interactions. Dev Dyn 218: 136-149. [Crossref]
- Wang T, Tamakoshi T, Uezato T, Shu F, Kanzaki-Kato N, et al. (2003) Forkhead transcription factor Foxf2 (LUN)-deficient mice exhibit abnormal development of secondary palate. Dev Biol 259: 83-94. [Crossref]
- Ormestad M, Astorga J, Landgren H, Wang T, Johansson BR, et al. (2006) Foxf1 and Foxf2 control murine gut development by limiting mesenchymal Wnt signaling and promoting extracellular matrix production. Development 133: 833-843. [Crossref]
- Nik AM, Johansson JA, Ghiami M, Reyahi A, Carlsson P (2016) Fo2021 Copyright OAT. All rights reservdevelopment and Tgfβ signaling in palatal shelf mesenchyme. Dev Biol 415: 14-23. [Crossref]
- Xu J, Liu H, Lan Y, Aronow BJ, Kalinichenko VV, et al. (2016) A Shh-Foxf-Fgf18-Shh Molecular Circuit Regulating Palate Development. PLoS Genet 12: e1005769. [Crossref]
- Nik AM, Reyahi A, Pontén F, Carlsson P (2013) Foxf2 in intestinal fibroblasts reduces numbers of Lgr5(+) stem cells and adenoma formation by inhibiting Wnt signaling. Gastroenterology 144: 1001-1011. [Crossref]
- Hirata H, Ueno K, Shahryari V, Deng G, Tanaka Y, et al. (2013) MicroRNA-182-5p promotes cell invasion and proliferation by down regulating FOXF2, RECK and MTSS1 genes in human prostate cancer. PLoS One 8: e55502. [Crossref]
- Kong PZ, Yang F, Li L, Li XQ, Feng YM (2013) Decreased FOXF2 mRNA expression indicates early-onset metastasis and poor prognosis for breast cancer patients with histological grade II tumor. PLoS One 8: e61591. [Crossref]
- Zhang Y, Wang X, Wang Z, Tang H, Fan H, et al. (2015) miR-182 promotes cell growth and invasion by targeting forkhead box F2 transcription factor in colorectal cancer. Oncol Rep 33: 2592-2598. [Crossref]
- Kundu ST, Byers LA, Peng DH, Roybal JD, Diao L, et al. (2016) The miR-200 family and the miR-183~96~182 cluster target Foxf2 to inhibit invasion and metastasis in lung cancers. Oncogene 35: 173-186. [Crossref]
- Shi Z, Liu J, Yu X, Huang J, Shen S, et al. (2016) Loss of FOXF2 Expression Predicts Poor Prognosis in Hepatocellular Carcinoma Patients. Ann Surg Oncol 23: 211-217. [Crossref]
- Wang QS, Kong PZ, Li XQ, Yang F, Feng YM (2015) FOXF2 deficiency promotes epithelial-mesenchymal transition and metastasis of basal-like breast cancer. Breast Cancer Res 17: 30. [Crossref]
- Zheng YZ, Wen J, Cao X, Yang H, Luo KJ, et al. (2015) Decreased mRNA expression of transcription factor forkhead box F2 is an indicator of poor prognosis in patients with resected esophageal squamous cell carcinoma. Mol Clin Oncol 3: 713-719. [Crossref]
- Tian HP, Lun SM, Huang HJ, He R, Kong PZ, et al. (2015) DNA Methylation Affects the SP1-regulated Transcription of FOXF2 in Breast Cancer Cells. J Biol Chem 290: 19173-19183. [Crossref]
- Lo PK, Lee JS, Liang X, Sukumar S (2016) The dual role of FOXF2 in regulation of DNA replication and the epithelial-mesenchymal transition in breast cancer progression. Cell Signal 28: 1502-1519. [Crossref]
- Milewski D, Pradhan A, Wang X, Cai Y, Le T, et al. (2017) FoxF1 and FoxF2 transcription factors synergistically promote rhabdomyosarcoma carcinogenesis by repressing transcription of p21(Cip1) CDK inhibitor. Oncogene 36: 850-862. [Crossref]
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, et al. (2006) A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10: 515-527. [Crossref]
- Riaz M, van Jaarsveld MT, Hollestelle A, Prager-van der Smissen WJ, Heine AA, et al. (2013) miRNA expression profiling of 51 human breast cancer cell lines reveals subtype and driver mutation-specific miRNAs. Breast Cancer Res 15: R33. [Crossref]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, et al. (2004) ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6: 1-6. [Crossref]
- Marcato P, Dean CA, Pan D, Araslanova R, Gillis M, et al. (2011) Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis. Stem Cells 29: 32-45. [Crossref]
- Marcato P, Dean CA, Liu RZ, Coyle KM, Bydoun M, et al. (2015) Aldehyde dehydrogenase 1A3 influences breast cancer progression via differential retinoic acid signaling. Mol Oncol 9: 17-31. [Crossref]
- Lewis MJ, Wiebe JP, Heathcote JG (2004) Expression of progesterone metabolizing enzyme genes (AKR1C1, AKR1C2, AKR1C3, SRD5A1, SRD5A2) is altered in human breast carcinoma. BMC Cancer 4: 27. [Crossref]
- Rybaczyk LA, Bashaw MJ, Pathak DR, Huang K (2008) An indicator of cancer: downregulation of monoamine oxidase-A in multiple organs and species. BMC Genomics 9: 134. [Crossref]
- Zareba I, Palka J (2016) Prolidase-proline dehydrogenase/proline oxidase-collagen biosynthesis axis as a potential interface of apoptosis/autophagy. Biofactors 42: 341-348. [Crossref]
- Luo W, Qin L, Li B, Liao Z, Liang J, et al. (2017) Inactivation of HMGCL promotes proliferation and metastasis of nasopharyngeal carcinoma by suppressing oxidative stress. Sci Rep 7: 11954. [Crossref]
- Cirillo F, Pellegrino M, Malivindi R, Rago V, Avino S, et al. (2017) GPER is involved in the regulation of the estrogen-metabolizing CYP1B1 enzyme in breast cancer. Oncotarget 8: 106608-106624. [Crossref]
- Gautam J, Banskota S, Regmi SC, Ahn S, Jeon YH, et al. (2016) Tryptophan hydroxylase 1 and 5-HT (7) receptor preferentially expressed in triple-negative breast cancer promote cancer progression through autocrine serotonin signaling. Mol Cancer 15: 75. [Crossref]
- Clendening JW, Pandyra A, Boutros PC, El Ghamrasni S, Khosravi F, et al. (2010) Dysregulation of the mevalonate pathway promotes transformation. Proc Natl Acad Sci USA 107: 15051-15056. [Crossref]
- Onono F, Subramanian T, Sunkara M, Subramanian KL, Spielmann HP, et al. (2013) Efficient use of exogenous isoprenols for protein isoprenylation by MDA-MB-231 cells is regulated independently of the mevalonate pathway. J Biol Chem 288: 27444-27455. [Crossref]





