Background and purpose: It has been suggested that consecutive DCE-MRI in the early phase of treatment with bevacizumab could provide a better estimate of treatment response.
Materials and methods: The xenograft study was performed using adult (8-12 weeks of age) male type NMRINUM Mice (Taconic, Denmark). A human glioblastoma xenograft was inoculated subcutaneously in the posterior flank of the mouse. DCE MRI and compartment analysis was performed extended Toft´s model. Pathology immunohistochemistry of Glial fibrillary acidic protein (GFAP) of excised tumors slides were analyzed using an ImageJ threshold watershed technique to determine tissue fraction of glial tumor cells.
Results: A total of 15 intervention tumors and two C16MG control tumors were subjected to DCE MRI. A significant correlation between Ktrans and GFAP was observed. Furthermore a significant reduction in Ktrans following Bevacizumab was observed.
Conclusion: To conclude this study successfully established a technique, which allowed small animals with Xenograft heterotopic implanted GBM tumors to be scanned with a DCE-MRI technique using a clinical 3T MR scanner setting. Furthermore the Ktrans parameters derived was demonstrated to be a potential imaging biomarker of GBM GFAP activity and treatment response to Bevacizumab.
glioblastoma, xenograft, bevacizumab, DCE-MRI, tumor
The majority of adult patients Glioblastoma die within 15–18 months from diagnosis, with less than 5% of patients alive at 5 years [1]. Regardless of age, patients should undergo a maximal safe resection and receive chemo-radiotherapy with Temozolomide [2]. High-grade gliomas are highly angiogenic and shown to secrete vascular endothelial growth factor A (VEGF-A), which acts in a paracrine manner to promote endothelial cell proliferation, survival and migration [3]. Accordingly, there is a convincing rationale for targeting the tumor vasculature though inhibition of the formation of new tumor blood vessels. Anti-angiogenic treatment may also cause normalization of existing tumor vasculature, which may lead to improved tumor drug delivery, and a synergistic effect of cytotoxic and antiangiogenic treatment [4].
Bevacizumab (Avastin, F. Hoffmann-La Roche, Basel, Switzerland) is a humanized monoclonal antibody that binds to and inhibits the activity of VEGF-A. In primary and recurrent glioblastoma, only Bevacizumab has so far shown meaningful efficacy in controlled clinical trials. However, the efficacy is limited to prolonging progression-free survival and to generating some additional palliative benefits, without affecting overall survival in the total population of glioblastoma patients [5]. GBM is a highly heterogeneous tumor that varies in mutation status, putative glial cell lineage, epigenetic profile and histological appearance [6]. This heterogeneity could explain why bevacizumab has a positive effect in only some patients. Further understanding of the mechanisms of resistance to bevacizumab treatment and better patient selection could improve outcomes for patients with GBM. Several biomarkers for better patient selection are being investigated, resulting among other findings in different molecular classifications that so far have not led to diversification in treatment [6]. Advanced magnetic resonance (MR) imaging such as dynamic contrast enhanced (DCE-MR) and diffusion-weighted (DWI-MR) are imaging techniques that can be used to acquire imaging features (imaging biomarkers), such as relative cerebral blood volume (rCBV), contrast volume transfer parameter (Ktrans), and the apparent diffusion coefficient (ADC) [7,8]. Several of these imaging biomarkers has been demonstrated to correlate to better treatment outcome, when treating malignant gliomas with antiangiogenic therapy [9-11]. It has been suggested that consecutive DCE-MRI in the early phase of treatment with bevacizumab could provide a better estimate of treatment response [12].
The purpose of the present study was to establish a technique of DCE-MRI in immunosuppressed mice with heterotopic implant of a GBM human tumor model using a clinical 3T MR scanner setting. In this clinical setup trying to establish correspondence between DCE-MRI perfusion and GBM activity estimated from GFAP staining of pathology tumor slides. Furthermore this study tried to estimate initial changes in DCE-MRI perfusion within days after treatment with bevacizumab in implanted tumors and to evaluate if DCE-MRI perfusion may serve as a surrogate biomarker of Bevacizumab treatment response.
Tumor model
The xenograft study was performed using adult (8-12 weeks of age) male type NMRINUM mice (Taconic, Denmark). A C16MG human glioblastoma xenografts growing in the immunosuppressed mice were used as tumor model. Tumors were initiated from established cell lines cultured in Dulbecco’s Modified Eagle´s Medium (DMEM-F12) with 10% fetal calf serum (FCS) and 1% penicillin/streptomycin added. Approximately 1 x 106 cells in 0.5 ml Matrigel were inoculated subcutaneously in the posterior flank of the mouse. Experiments were initiated when the tumors had either grown to a volume of >200 mm3 or shown signs of infiltrative growth (day zero).
Animal procedure
MR imaging was conducted on anesthetized mice. The mice were anesthetized with Hypnorm (fentanyl citrate 0.315 mg/ml and fluanisol 10 mg/ml) and Midazolam (5 mg/ml). The two drugs were mixed with sterile water with the ratio two parts sterile water, one part Hypnorm and one part Midazolam. The dose was 0.01 ml/g mouse. On day one of the experiment, the tumors were scanned by MRI (scan 1). Immediately after the scan, the tumor was treated with 10 mg/kg Bevacizumab administered intra peritoneal. For each ten tumors treated with Bevacizumab (intervention tumors), one tumor was treated with 0.9% saline (control tumor). Each tumor was scanned again on day three (scan 2) and day eight (scan 3). Immediately after the scan on day eight, the tumor was given a second Bevacizumab (intervention tumor) or saline (control tumor) injection. The last scan was performed on day ten (scan 4). Dotarem (Guerbet, Aulnay-sous-Bois, France), diluted in 0.9% saline to a final concentration of 56 μmol/ml, was used as the contrast agent. The contrast agent was administered in the tail vein of the mice in a bolus dose of 0.1 mmol/kg. The contrast was injected into the tail vein of the mouse through a 25G-needle attached to an 80 cm fine-bore polythene tube.
MR imaging
MRI was performed using a 3.0-T whole-body scanner (Ingenia 3T, Philips Healthcare, Best, the Netherlands) and a single-element microscopy coil with an inner diameter of 47 mm (dS Microscopy, Philips Healthcare). The microscopy coil was placed in an in-house fabricated bed consisting of three different Styrofoam plates (Figure 1). The bottom plate was a 40 mm thick bottom plate with a milled groove holding a 500 ml saline bag (58⁰C), which kept the temperature in the bed stable at 32⁰C during the scan. The middle plate was an 80 mm thick middle plate with a milled groove for the mouse container and microscopy coil, including holes for the oxygen supply (2 l/min). The top plate was an 80 mm thick top plate to close the bed. The bed was fixed to the scanner couch to ensure that the mice were placed in the iso-center of the scanner. The temperature of 32⁰C in the bed kept the body core of the mice at a stable temperature between 37⁰C and 38⁰C during the scan. Further detailed Information about the development of the mouse bed is intended for publication elsewhere [13].

Figure 1. Photo of mice in in-house made scanning phantom. The animal is in Plexiglas tube with tail fixated. A 24 gauge Venflon (Pediatric) for contrast administration has been inserted in tail vein. Below the animal small heat vents allow air heated by the hot water reservoir below the animal to flow upwards to keep the animal warm. In front og the animal a tube allowing oxygen to flow to keep the animal well oxygenated once a polystyrene slab is put in top and the animal is enclosed during scanning. The micro coil are placed over the tumor bearing flank area. The phantom is fixated to the couch of the MR scanner to ensure the same position in the bore for each scan
The scan protocol consisted of T2-weighted (T2W) axial images for tumor anatomy and further Planning, and a DCE-MRI protocol consisting of the following 3 scan sequences: a 3D T1 fast field Echo pre-scan sequence with TR = 50 ms, TE = 2.5 ms, flip angle (FA) = 6⁰, FOV = 50 x 40 mm2, Matrix 84 x 67 pixels, slice thickness = 0.6 mm (corresponding to a voxel size of 0.6 x 0.6 x 0.6 mm3), and an inter slice gap = 0 mm. Interpolation algorithms applied by the imaging system resulted in an Apparent resolution of 0.20 x 0.19 x 0.60 mm3. This sequence was followed by an identical pre-scan Sequence, but with a flip angle of 16⁰. Finally, a dynamic 3D T1 turbo field echo sequence with TR = 6.7 ms, TE = 2.5 ms, the same voxel size and inter slice gap as in the pre-scans, and 12⁰ flip angle was used during the contrast infusion. Volume images were sampled at 5.4-second intervals for a total of 80 dynamic scans. The last sequence consisted of a 3D T1 axial turbo field echo sequence with TR = 25 ms, TE = 12 ms and voxel size 0.3 mm3 (recon voxel size 0.09 x 0.09 x 0.30 mm3). This scan was used to obtain a high-resolution post-contrast image of the tumor anatomy.
Compartment analysis was performed using the MR perfusion tool in the IntelliSpace Portal System (IPS, Version 5; Philips), which uses extended Toft´s model [14]. One of the mouse´s iliac vessels (Figure 2A), which could be identified in each scan with good reproducibility, was chosen as an input function for the analysis. Ktrans was used as a biomarker of tumor angiogenesis. A volume of interest (VOI) covering the xenograft tumor was contoured on all slices with visible tumor. The VOI was contoured with a margin of approximately 1 mm to the skin surface to avoid Ktrans map artifacts. For a tumor that infiltrated the underlying muscle beyond the implantation site, the VOI was drawn to include only tumor outside the muscle. Contouring was performed on the Ktrans map with the T2W images as background (Figure 2C+2D). The contour was checked against the dynamic T1W frame with maximal contrast enhancement (Figure 2B). All four DCE-MRI scans were contoured for each animal. Finally, the mean Ktrans value for each VOI was calculated and used as the imaging biomarker.
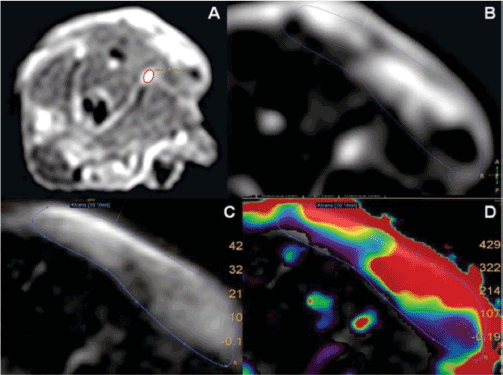
Figure 2. Axial DCE-MRI scan of a mouse placed on its stomach with its head in the head direction of the scanner with no supination/pronation correction added, resulting in orientation og image right representing mouse right. A: Right iliac vessel (red ROI) chosen as input function for DCE-MRI data analysis B: Dynamic T1W frame with maximal contrast enhancement C: Gray-scaled Ktrans map with T2W images as background D: Color-scaled Ktrans map with T2W images as background.
Blue ROI = tumor contour
Tumor volumes were measured as a 3D volume from MR scans using the IntelliSpace Portal System (IPS, Version 5; Philips).
Pathology
Each animal was euthanized immediately after the last scan (day 10), and the tumor excised and fixed to a cork plate by pins to avoid the skin to curl up. Fixation was in 4 per cent formalin. The whole tumor was sectioned perpendicular to the skin surface. The tissue blocks including normal skin and the whole tumor were paraffin-embedded. From the paraffin-embedded tissue blocks sections were cut for staining. Tumor immunohistochemistry contained staining for the protein cluster of differentiation 31 (CD31), which in humans is encoded by the PECAM-1. CD31 primarily demonstrate the presence of endothelial cells in histological tissue sections to evaluate the degree of tumor angiogenesis. Glial fibrillary acidic protein (GFAP), an intermediate filament protein, expressed by numerous cell types of the central nervous system (CNS) including astrocytes. GFAP was used to identify GBM tumor cells. Finally, Hematoxylin and eosin stain (HE) was applied to identify collagen structures. All immunohistochemistry was performed within the same antibody batch.
Pathological slides were scanned on a Hamamatsu scanner. Slide images were exported as tiff files using the free software tool NDP view version 2.6.13 from Hamamatsu. Implanted tumors were outlined manually on each pathology slide image. A small program (macro) was written for each image using ImageJ [15]. Each macro for analyzing a specific slide was saved for later test of the best threshold in a reproducible way. ImageJ outlined images were converted to 8 bit greyscale images. Images were subsequently made binary (black and white images) using an overall greyscale threshold value of 120 on all images. The chosen threshold value ensured stable results even with small changes in threshold value, and gave reasonable results when overlaid the original pathology images. A watershed method was used to identify black particles (stained cells) size 100 pixels or larger. The values of 100 pixels is again empirically determined based on robust outcome of the analysis. Finally the stained fraction in percentages (GFAP %) within the outlined tumor area was calculated.
A total of 15 C16MG intervention tumors and two C16MG control tumors were subjected to MRI.
Measured values of tumor volume, Ktrans and GFAP% from each day of MR scanning are shown in Table 1. One animal dies before the second Bevacizumab injection (animal 12). The Ktrans of the two control tumors continued to rise throughout the entire observation period, i.e. no effect was seen following placebo treatment.
Table 1. The table gives the tumor volume and the mean Ktrans value for each tumor volume for each day of MR scanning. On day 10 animals were euthanized and their tumors excised. Subsequently pathology slides were stained with GFAP and the percentage positive for GFAP estimated (GFAP%). Ktrans was estimated immediately before each of the two days of Bevacizumab treatment Ktrans(1) and Ktrans(8) ( Ktrans on day 1 and 8), and Ktrans two days after each treatment Ktrans(3) and Ktrans(10) (Ktrans on day 3 and 10). Animals C1 and C2 were not treated with Bevacizumab. * = Animal no 12 died before the second Bevacizumab injection
Animal
No |
Day_1 |
Day_3 |
Day_8 |
Day_10 |
Volume |
Ktrans(1) |
Volume |
Ktrans(3) |
Volume |
Ktrans(8) |
Volume |
Ktrans(10) |
GFAP% |
mm3 |
10-3 min-1 |
mm3 |
10-3 min-1 |
mm3 |
10-3 min-1 |
mm3 |
10-3 min-1 |
% |
1 |
33 |
217 |
39 |
155 |
73 |
190 |
90 |
142 |
6 |
2 |
60 |
897 |
57 |
616 |
31 |
375 |
56 |
377 |
43 |
3 |
296 |
170 |
214 |
101 |
180 |
150 |
217 |
205 |
8 |
4 |
35 |
583 |
31 |
150 |
21 |
203 |
34 |
410 |
30 |
5 |
247 |
309 |
192 |
201 |
208 |
498 |
115 |
588 |
13 |
6 |
60 |
395 |
60 |
266 |
60 |
413 |
60 |
201 |
7 |
7 |
84 |
525 |
93 |
262 |
119 |
409 |
65 |
398 |
14 |
8 |
75 |
367 |
124 |
205 |
56 |
647 |
57 |
222 |
8 |
9 |
179 |
117 |
177 |
167 |
404 |
395 |
269 |
356 |
1 |
10 |
143 |
341 |
180 |
262 |
153 |
364 |
70 |
244 |
9 |
11 |
84 |
1063 |
63 |
179 |
93 |
313 |
116 |
216 |
3 |
12* |
256 |
115 |
249 |
81 |
missing |
|
missing |
|
missing |
13 |
79 |
188 |
126 |
133 |
126 |
145 |
172 |
136 |
22 |
14 |
59 |
359 |
67 |
264 |
90 |
143 |
98 |
192 |
7 |
15 |
380 |
238 |
129 |
745 |
124 |
689 |
192 |
176 |
14 |
|
|
|
|
|
|
|
|
|
|
C1 |
73 |
103 |
64 |
298 |
75 |
478 |
76 |
569 |
34 |
C2 |
40 |
140 |
65 |
350 |
54 |
499 |
60 |
565 |
24 |
Mean |
128 |
360 |
114 |
261 |
110 |
369 |
103 |
312 |
14 |
StdDev |
102 |
266 |
65 |
170 |
90 |
166 |
66 |
152 |
12 |
For all tumors treated with Bevacizumab, CD31 staining of the pathology slides gave very sparse signal indicating effect of Bevacizumab as shown for the tumor in animal 15 on Figure 3A. Consequently, segmentation of CD31 was not possible in pathology slides from tumors of treated animals. On the contrary, the CD31 staining of the tumors in the control animals demonstrated a positive staining with CD31 as seen on Figure 3D.
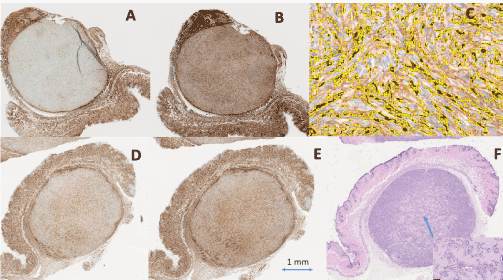
Figure 3. Pathology immunohistochemistry slides from animal No. 15 (treated with Bevacizumab). A) Tumor stained with CD31 B) Tumor stained with GFAP C) Enlarged section of B) demonstrating the watershed segmentation of stained particles. Pathology immunohistochemistry slides from animal C2 (treated with placebo) D) Tumor stained with CD31 E) Tumor stained with GFAP F) Tumor stained with Hematoxylin and Eosin. Scale of all pathology images are shown in image E). The exact scale could not be accessed for the watershed image C), as the image was obtained as a screen dump
Tumors in both treated, Figure 3B, and control animals, Figure 3E, demonstrated positive immunochemistry staining with GFAP, and segmentation of GFAP was possible in all tumors. One example of the watershed segmentation is shown in Figure 3C. One example of Hematoxylin and Eosin staining, used to demonstrate the existence of collagen after treatment, is shown in Figure 3F.
The percentage positive GFAP stain (GFAP %) was plotted against the corresponding Ktrans values from day 10 in Figure 4, and a linear regression demonstrated the expected positive correlation: Regression line: GFAP% (Day 10) = 0.040 * Ktrans (Day 10) + 2,94 with a correlation coefficient = 0.51. T-test of the slope was significant with p=0.04. No significant relation between GFAP % and tumor volume at day 10 could be established (data not shown).
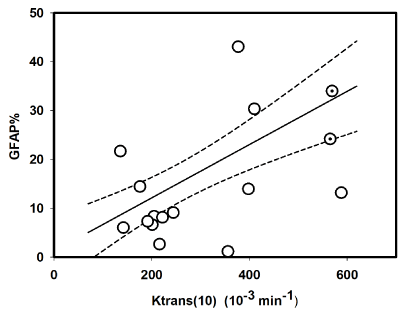
Figure 4. White circles represent plot of GFAP percentage estimated from histopathology GFAP slides versus Ktrans on day 10 (day of euthanasia of animal) for fourteen of the fifteen animals in the experiment (one animal died before end of experiment). The two dotted white circles represent values for the two untreated animals. The plot also show the regression line (solid) and corresponding 95 % confidence bands (dotted). Regression analysis results were: GFAP % (Day 10) = 0.040 * Ktrans (Day 10) + 2,94 with a correlation coefficient = 0.51. T-test of the slope was significant with p=0.04
Paired values of Ktrans and tumor volumes could be determined in all four scanning days. After logaritimic transformation a significant correlation could be established as shown in Figure 5. Regression line: log (Ktrans) = -0.28 * log (tumor volume) + 3.00 with a correlation coefficient = 0.30. Intercept and slope t-test was significant with p<0.0001 and p=0.015 respectively. Subsequently Ktrans values were standardized to the same tumor volume using the established relation between Ktrans and tumor volume. Using the standardized Ktrans values regression analysis demonstrated an improved correlation compared to using uncorrected Ktrans values: GFAP% = 0.05 * Ktrans (10) std + 1.59 with a correlation coefficient of 0.63. The slope was statistically significant with p = 0.008. The standardized Ktrans values are given in table 2 for each MR scanning. The Ktrans values determined 2 days after the first Bevacizumab treatment was significant lower than corresponding pre-treatment values (Wilcoxon Matched Pairs Test p-value = 0,012). The relative change in Ktrans following each Bevacizumab treatment injection was calculated as the ratio between Ktrans after and before and treatment: SF1 = Ktrans (Day 3)_std / Ktrans (Day 1)_std and SF2 = Ktrans (Day 10)_std / Ktrans (Day 8)_std. Results are shown in table 2. Of the 15 animals treated with Bevacizumab, 13 tumors showed an initial response in terms of a reduction in Ktrans two days after the first Bevacizumab injection. Two tumors demonstrated an initial increase in Ktrans (animal 9 and 15), the tumor in animal no.15 later responds to the second Bevacizumab injection. A paired t-test between logarithmic values of SF1 and SF2 demonstrated non-significant trend (Mean SF1 = 0.66 and SF2 = 0.81, p = 0.42). SF1 in animal no. 15, however presented as an extreme outlier. If animal no.15 was treated as an outlier and left out, the previous trend become significant (Mean SF1 = 0.57 and SF2 = 0.89, p = 0.02).
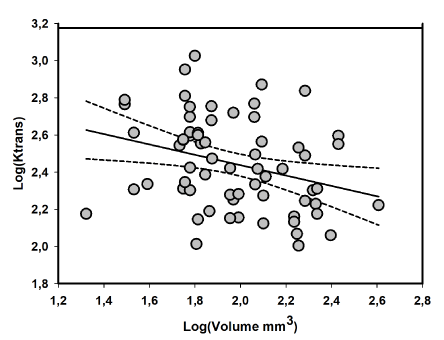
Figure 5. Grey circles represent plot of corresponding logarithmic values of Ktrans versus implanted tumor volume on each DCE MR scanning day (Day 1,3,8 and 10). Included in the plot are also the regression line (solid) and corresponding 95 % confidence bands (dotted). Regression analysis results were: log (Ktrans) = -0.28 * log (tumor volume) + 3.00 with a correlation coefficient = 0.30. T-test of the slope was with p = 0.015
Table 2. The table gives Ktrans standardized values (Ktrans_std) corrected for difference in tumor volume corresponding to the original values in table 1. SF1 and SF2 are relative change in Ktrans_std from day 1 to day 3 and day 8 to day 10 respectively
Animal
No |
Day_1 |
Day_3 |
Day_8 |
Day_10 |
SF1 |
SF2 |
Ktran_std |
Ktran_std |
Ktran_std |
Ktran_std |
10-3 min-1 |
10-3 min-1 |
10-3 min-1 |
10-3 min-1 |
1 |
244 |
166 |
171 |
121 |
0.68 |
0.70 |
2 |
853 |
594 |
429 |
365 |
0.70 |
0.85 |
3 |
103 |
67 |
105 |
136 |
0.65 |
1.30 |
4 |
644 |
171 |
259 |
457 |
0.27 |
1.77 |
5 |
198 |
138 |
335 |
466 |
0.70 |
1.39 |
6 |
375 |
253 |
393 |
191 |
0.67 |
0.49 |
7 |
454 |
220 |
321 |
370 |
0.49 |
1.15 |
8 |
328 |
159 |
627 |
214 |
0.49 |
0.34 |
9 |
82 |
117 |
221 |
223 |
1.43 |
1.01 |
10 |
254 |
183 |
266 |
222 |
0.72 |
0.83 |
11 |
920 |
168 |
263 |
171 |
0.18 |
0.65 |
12* |
73 |
52 |
missing |
missing |
0.71 |
missing |
13 |
165 |
103 |
112 |
96 |
0.62 |
0.86 |
14 |
343 |
244 |
122 |
159 |
0.71 |
1.31 |
15 |
135 |
572 |
535 |
121 |
4.23 |
0.23 |
C1 |
93 |
278 |
427 |
506 |
NA |
NA |
C2 |
149 |
326 |
488 |
537 |
NA |
NA |
Mean |
318 |
224 |
298 |
256 |
0.8 |
0.8 |
StdDev |
254 |
149 |
152 |
148 |
0.9 |
0.4 |
The present study successfully established a technique, which allowed small animals with Xenograft tumors to be scanned using a clinical 3T MR scanner setting. Images obtained on the clinical MR scanner will look more blurred than similar images from an experimental animal scanner, this is mainly due to the lower magnetic field strength the clinical MR scanner compared to small animal MR scanners which typically have magnetic field strengths of 9-15T. Blurring was also the reason for using a heterotopic flank implantation of the GBM tumors, which allowed tumors to grow to a larger size than in the brain before euthanizing the animals.
The images obtained in the present study on the clinical MR scanner actually demonstrated a significant correlation between Ktrans from DCE-MRI perfusion and GBM activity estimated from GFAP staining of pathology tumor slides, thus demonstrating clinically estimated Ktrans may be a valid imaging biomarker of GBM activity.
Unexpectedly, a significant correlation between Ktrans and tumor volume could not be demonstrated in the present study. This observation may be explained from the fact that Hematoxylin Eosin stain demonstrated the large formations of collagen in this human GBM tumor line. Variable collagen formation in the implanted tumors may be a source of the relative large variation around the regression line in Figure 3, which is also supported by an improved correlation to GFAP% following the correction of Ktrans for volume dependency.
Even though a limitation in the present study, the use of using only one human tumor line was justified as this study was a proof of concept for using a clinical MR scanner for small animals.
Tumor histopathology two days after the last Bevacizumab injection demonstrated sparse endothelial tissue. A similar result has been observed in another study that showed increased vascular mimicry and negative staining for CD31 three days after Bevacizumab treatment [16]. Vascular mimicry may be an explaining factor of the observation of negative CD31 staining despite of increasing Ktrans two days after the last Bevacizumab injection in all treated animals, while positive CD31 staining was observed in the control animals.
Initially, a statistically significant decrease in Ktrans from pre- to post-treatment values were observed, indicating that change in Ktrans may be a biomarker of response to the tumors vascular system. Similar results have been observed in other studies [17]. When the relative change in Ktrans after the first Bevacizumab injection SF1 was compared to the analogous value after the second treatment SF2, a non-significant trend towards resistance, decreased effect from bevacizumab treatment. If one animal with an extreme outlier value of SF1 was omitted from analysis, the trend of a reduction in Bevacizumab treatment effect became statistically significant. In previous studies, we observed that tumor drug sensitivity could be represented by a spectral distribution [18]. So the results in the present study may lead to an interesting new hypothesis: that of spectral anti-angiogenesis activity in this human GBM tumor line. This observation is supported by recent observations of anti-angiogenic therapy being quite complex with several different mechanisms of resistance have been described [19]. Further studies, both pre-clinical and clinical, will be necessary to validate these findings and this new hypothesis from the present study.
To conclude this study successfully established a technique, which allowed small animals with Xenograft heterotopic implanted GBM tumors to be scanned with a DCE-MRI technique using a clinical 3T MR scanner setting. Furthermore the Ktrans parameters was demonstrated to be a potential imaging biomarker of GBM GFAP activity and treatment response to Bevacizumab.
Torben Moos, Department of Health Science and Technology, Aalborg University for donating the GBM tumor cells Benedict Kjærgaard, Department of Clinical Medicine, Aalborg University Hospital for providing and taking care of our experimental animals Dennis Tideman Arp and Kristian Lund, Department of Medical Physics, Oncology, Aalborg University Hospital for help with MR scanning.
No external funding was received for the present project. None of the authors have any conflicts of interest to declare and are solely responsible for the present publication. National, and institutional guidelines for the care and use of animals were followed. The animal care and experimental procedures were approved by the Danish Animal Experiments Inspectorate, license number 2014-15-0201-0043.
- Ostrom QT, Gittleman H, Farah P, Ondracek A, Chen Y, et al. (2013) CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006-2010. Neuro Oncol 15: 1-56.
- Wick W, Osswald M, Wick A, Winkler F (2018) Treatment of glioblastoma in adults. Ther Adv Neurol Disord 11: 1756286418790452.
- Millauer B, Shawver LK, Plate KH, Risau W, Ullrich A (1994) Glioblastoma growth inhibited in vivo by a dominant-negative Flk-1 mutant. Nature 367: 576-579.
- Dickson PV, Hamner JB, Sims TL, Fraga CH, Ng CY, et al. (2007) Bevacizumab-induced transient remodeling of the vasculature in neuroblastoma xenografts results in improved delivery and efficacy of systemically administered chemotherapy. Clin Cancer Res 13: 3942-3950.
- Winkler F, Osswald M, Wick W (2018) Anti-Angiogenics: Their Role in the Treatment of Glioblastoma. Oncol Res Treat 41: 181-186.
- Keunen O, Taxt T, Gruner R, Lund-Johansen M, Tonn JC, et al. (2014) Multimodal imaging of gliomas in the context of evolving cellular and molecular therapies. Adv Drug Deliv Rev 76: 98-115.
- Daniels D, Guez D, Last D, Hoffmann C, Nass D, et al. (2016) Early Biomarkers from Conventional and Delayed-Contrast MRI to Predict the Response to Bevacizumab in Recurrent High-Grade Gliomas. Am J Neuroradiol 37: 2003-2009.
- Santos P, Peck KK, Arevalo-Perez J, Karimi S, Lis E, et al. (2017) T1-Weighted Dynamic Contrast-Enhanced MR Perfusion Imaging Characterizes Tumor Response to Radiation Therapy in Chordoma. Am J Neuroradiol 38: 2210-2216.
- Kong Z, Yan C, Zhu R, Wang J, Wang Y, et al. Imaging biomarkers guided anti-angiogenic therapy for malignant gliomas. Neuroimage Clin 20: 51-60.
- Ellingson BM, Cloughesy TF, Lai A, Mischel PS, Nghiemphu PL, et al. (2011) Graded functional diffusion map-defined characteristics of apparent diffusion coefficients predict overall survival in recurrent glioblastoma treated with bevacizumab. Neuro Oncol 13: 1151-1161.
- Kim R, Choi SH, Yun TJ, Lee ST, Park CK, et al. (2017) Prognosis prediction of non-enhancing T2 high signal intensity lesions in glioblastoma patients after standard treatment: application of dynamic contrast-enhanced MR imaging. Eur Radiol 27: 1176-1185.
- Kickingereder P, Wiestler B, Graf M, Heiland S, Schlemmer HP, et al. (2015) Evaluation of dynamic contrast-enhanced MRI derived microvascular permeability in recurrent glioblastoma treated with bevacizumab. J Neurooncol 121: 373-380.
- Ravn S, Arp DT, Lund K, Magnusdottir SO, Kjærgaard B, et al. (2019) Dynamic contrast-enhanced magnetic resonance imaging of Glioblastoma Multiforme response to Bevacizumab - a xenograft study.
- Tofts PS, Brix G, Buckley DL, Evelhoch JL, Henderson E, et al. (1999) Estimating kinetic parameters from dynamic contrast-enhanced T(1)-weighted MRI of a diffusable tracer: standardized quantities and symbols. J Magn Reson Imaging 10: 223-232.
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671-675.
- Xue W, Du X, Wu H, Liu H, Xie T, et al. (2017) Aberrant glioblastoma neovascularization patterns and their correlation with DCE-MRI-derived parameters following temozolomide and bevacizumab treatment. Sci Rep 7: 13894.
- O'Neill AF, Qin L, Wen PY, de Groot JF, Van den Abbeele AD, et al. (2016) Demonstration of DCE-MRI as an early pharmacodynamic biomarker of response to VEGF Trap in glioblastoma. J Neurooncol 130: 495-503.
- Carl J (1989) Drug-resistance patterns assessed from tumor marker analysis. J Natl Cancer Inst 81: 1631-1639.
- Pezzella F (2019) Mechanisms of resistance to anti-angiogenic treatments. Cancer Drug Resistance 2: 595-607.





