Abstract
Objective: Memantine, a low-affinity N-methyl-D-aspartate receptor antagonist, is one of the primary pharmacological therapies for Alzheimer’s disease. To explore novel actions of memantine, we analyzed effects of memantine on cell viability, cell growth, and protein profile of neuroblastoma cells. Methods: A human neuroblastoma cell line of GIMEN was used. GIMEN cells were cultured in the presence or absence of 1-100 mM memantine for 48 hours. Effects of memantine on the cell viability and growth were evaluated by counting cell numbers. Proteins, extracted from GIMEN cells treated or non-treated with 10 mM memantine, were separated by 2 dimensional-differential image gel electrophoresis (2D-DIGE). Protein spots of interest were subjected to protein identification by mass spectrometry. Results: The viability of GIMEN cells was 99.0% or more in the range of 1-10 mM memantine compared to that in the absence of memantine. The viability was slightly decreased in the presence of 100 mM memantine (97.5%, p<0.01). In contrast, the cell growth was suppressed by memantine in a dose-dependent manner (2-100 mM, 85.5-63.0%; 2-10 mM, p<0.05; 20-100 mM, p<0.01). For the protein profile analysis, we used GIMEN cells treated with 10 mM memantine which showed the viability of 99.4% and the growth of 76.1%. As a result, 892 protein spots were detected in the 2D-DIGE results of 10 mM memantine-treated and non-treated GIMEN cells. 13 protein spots showed 1.2-fold or higher intensity and 19 protein spots showed -1.2 (1/1.2)-fold or lower intensity in the memantine-treated cells than in the non-treated cells (p<0.05). We identified proteins in 7 out of the 32 spots: coronin-1C (1.44-fold increased), β-actin (-1.21-fold decreased), g-enolase (-1.21-fold decreased), glutathione synthetase (-1.28-fold decreased), spermatogenesis-associated protein 24 (-1.46-fold decreased), and V-set transmembrane domain-containing protein 2B (-1.46-fold decreased). Interestingly, β-actin, g-enolase, and glutathione synthetase are known to be involved in cell proliferation. Conclusion: Memantine suppressed the growth of GIMEN cells and affected their protein profiles. Our data suggested a novel action of memantine to suppress the neuroblastoma cell growth, which may be associated with the decreased expression of β-actin, g-enolase, and glutathione synthetase.
Key words
Alzheimer’s disease, cell growth, mass spectrometry, memantine, neuroblastoma cells, protein profiles
Introduction
Alzheimer’s disease (AD), a neurodegenerative disorder, is the main cause of dementia [1, 2]. Although the etiology of AD remains obscure, it is believed to be associated with the formation of senile plaques and neurofibrillary tangles in the brain due to the extracellular deposition of amyloid β (Aβ) peptides and hyperphosphorylation of intracellular tau protein [2]. Vaccines or antibodies targeting these aberrant peptides/protein and inhibitors for β-secretase or g-secretase, which interfere with the production of amyloid β peptides, are under investigation for treatment of AD [3]. However, none of these drugs has brought a fundamental breakthrough [4]. Choline esterase inhibitors and N-methyl D-aspartate (NMDA) receptor antagonist are currently the main treatment for AD, the latter of which has a unique neuroprotective effect [4, 5].
NMDA receptors, one of ion channel-type glutamate receptors, are crucial for neuronal communication [6]. To date, 7 different subunits of NMDA receptors have been identified: NR1, NR2A-D, and NR3A and B [6, 7]. NMDA receptors are heterotetramers that consist of two NR1 subunits and two NR2 subunits or two NR1 subunits and a mixture of NR2 and NR3 subunits [6]. Glycine and glutamate bind to NR1 and NR2, respectively, and the glycine sites must be occupied before the glutamate-binding for the activation of the receptors [7]. NMDA receptors that are non-selective cation channels have a potential for high Ca2+ permeability [6]. At rest, Mg2+ binds to NMDA receptors to block the activation. On sufficient depolarization of the synaptic membrane, Mg2+ dissociates from NMDA receptors in a voltage-dependent manner [6, 7]. Simultaneously, glutamate must bind to the receptors for their activation. The activation of NMDA receptors opens their ion channels for Ca2+ influx to mediate long-term potentiation (LTP) [6, 8]. LTP has been implicated to play a crucial role in the processes of learning and memory formation [8].
In the brains of AD patients, Aβ peptides mediate an increase in extracellular glutamate by increasing glutamate release from presynaptic neurons, induction of glutamate release from astrocytes and microglia, and inhibition of glutamate uptake by astrocytes and microglia [9, 10]. Prolonged activation of synaptic NMDA receptors and another ion channel-type glutamate receptor, α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid (AMPA) receptors, leads to their desensitization and internalization, resulting in synaptic depression. Finally, excess glutamate activates extra-synaptic NMDA receptors, which results in multiple deleterious downstream events, including cell death and impairment of LTP [6, 9]. Hyperphosphorylated tau targets the receptor protein kinase Fyn that phosphorylates NR2B to stabilize their association with postsynaptic density 95 (PSD95), which enhances the above glutamatergic excitotoxicity induced by Aβ peptides.
Memantine is a low affinity, voltage-dependent, non-competitive NMDA receptor antagonist which is currently used in the treatment of moderate to severe AD [6, 11, 12]. Compared to other NMDA receptor channel blockers, memantine has a higher affinity than that of Mg2+, but a lower affinity than those of ketamine, phencyclidine, and MK-801 [13]. Memantine enters NMDA receptor channel preferentially when the channel is excessively open [14]. Thereby, memantine inhibits the Ca2+ influx by blocking the channel activity that is induced by relatively low, tonic levels of glutamate [11-13]. This may contribute to symptomatic improvement of AD and protect neuronal cells from the glutamatergic excitotoxicity [12, 13]. Most importantly, the off-rate of memantine from NMDA receptors is relatively fast, which allows for the preservation of the physiological function of the receptors including expression of LTP [11, 14]. NMDA receptors blocked by memantine can be activated by higher concentrations of glutamate released after depolarization of the presynaptic neurons [11, 13].
Besides the above-mentioned action as an NMDA receptor antagonist, memantine has been found to influence protein expression of neuronal cells. Memantine reduces levels of total Aβ precursor protein (APP), APPα isoform, and Aβ1-40 secreted from neuroblastoma cells, and lowers the amyloidogenic Aβ1-42 secretion in primary rat cortical neuronal culture [15, 16]. Memantine also inhibits and reverses protein phosphatase 2A inhibition-induced abnormal hyperphosphorylation and accumulation of tau in organotypic culture of rat hippocampal slices [17]. Furthermore, memantine affects physiologically expressed genes/proteins in rat and mouse brains [18, 19]. However, no comprehensive study of physiologically expressed proteins in human neuronal cells under the effect of memantine has been conducted to date as far as we searched. It is important to collect information concerning the molecular pharmacology of memantine to understand its actions and anticipate potential adverse effects.
In this study, we analyzed effects of memantine on the viability, growth, and protein profile of neuroblastoma cells. We found that memantine almost did not affect the cell viability but decreased the cell growth in a dose-dependent manner. Protein profile of neuroblastoma cells was changed by memantine, showing altered expression levels of at least 6 proteins. Our findings suggest novel actions of memantine, which may underlie the success of AD treatment.
Materials and methods
Cell culture
A human neuroblastoma cell line of GIMEN (CLS Cell Lines Service GmbH, Eppelheim, Germany) was used in this study. GIMEN cells were cultured in DMEM supplemented with 10% FBS. To investigate effects of memantine on the cell viability and proliferation, GIMEN cells were cultured in the presence or absence of 1-100 mM memantine for 48 hours. Specifically, the cells were plated onto a 96-well plate at 7.0×104 cells/cm2. Then, the culture medium was replaced with FBS-free DMEM that contained or did not contain 1-100 mM memantine. After culturing for 48 hours, the cell viability was calculated as ratios of the viable cell numbers to the total cell numbers. The cell growth was calculated as ratios of the viable cell numbers (at 48 hours) to the seeded cell numbers (2.2 x 104 cells, at 0 hour). To perform 2-dimensional differential image gel electrophoresis (2D-DIGE), cytokine array, and ELISA, the cells were cultured for 48 hours in the presence or absence of 10 mM memantine. All the experiments were performed in triplicates.
Reverse-transcription polymerase chain reaction (RT-PCR)
RNA extraction and RT-PCR were performed using TaqMan Gene Expression Cells-to-CT Kit (Thermo Fisher Scientific, Waltham, MA, USA) and ExTaq (Takara Bio Inc., Shiga, Japan). Sequences of primers for NMDA receptor subunits and 18S ribosomal RNA were previously reported [20]. PCR products were separated by 3% agarose gel electrophoresis.
2D-DIGE
Proteins extracted from GIMEN cells were separated by 2D-DIGE as described previously [21]. Briefly, an equal weight of proteins taken from six lysates (three of 10 μM memantine-treated and three non-treated) were mixed and labeled with Cyanine dye 3 (Cy3, Cy Dye DIGE Saturation dye, GE Healthcare, Buckinghamshire, UK) for preparation of an internal control “standard sample”. Each of the six samples was labeled with Cyanine dye 5 (Cy5, GE Healthcare). 2.5 mg of the individual Cy5-labeled samples were mixed with 2.5 mg of the Cy3-labeled standard sample. Then each of the mixed protein sample was applied to an isoelectric focusing (IEF) gel strip (Immobiline Drystrip pH 3-11, 24 cm, non-linear, GE Healthcare). After the proteins were separated by IEF, they were further separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The resultant protein spots were scanned using an image analyzer (Typhoon 9400 Imager, GE Healthcare). The Cy5-fluorescent intensity of each protein spot was normalized by the Cy3-fluorescent intensity of an identical spot by using a quantitative analysis program (Progenesis, Nonlinear Dynamics, Newcastle, UK). The normalized Cy5-fluorescent intensity was compared between the memantine-treated and non-treated samples using Progenesis.
Protein identification
Proteins were identified by mass spectrometry (MS) [21]. In brief, 50 mg of proteins was separated by 2-dimensional electrophoresis. Gel specimens corresponding to protein spots of interest were recovered. Then proteins in the gel fragments were digested with trypsin. Peptides produced by the digestion were analyzed using a matrix-assisted laser desorption/ionization time of flight mass spectrometer (MALDI-TOF/MS) (Ultraflex, Bruker Daltonics, Ettlingen, Germany). Based on the mass spectra, some peptides were selected for MS/MS analysis. The obtained MS and MS/MS spectra were used to identify the proteins by database searching (Mascot, http://www.matrixscience.com) against the Swiss Prot human protein sequence database. Protein identification was accepted when MASCOT search results delivered significant MOWSE scores (p<0.05).
Analysis of the secretion of humoral factors from GIMEN cells treated with memantine
Humoral factors in culture supernatants were measured in duplicate by a cytokine array (Human Neuro Discovery Array C1, RayBiotech, Norcross, GA, USA). The analyzed humoral factors were as follows: BDNF, βNGF, GCSF, GDNF, HB-EGF, IFNg, IGF-1, IL-10, IL-1α, IL-1β, IL-6, IL-8, MCP-1, MIP-1α, MMP-2, MMP-3, S100B, TGFβ, TNFα, and VEGF-A. Humoral factors detected as specific spots were visualized using an image analyzer (LAS-3000, Fujifilm, Tokyo, Japan). The intensity of each spot, which was measured by Science Lab 2003 Image Gauge software program (Fujifilm), was normalized based on the intensity of positive control spots. MCP-1, a chemokine selected by the cytokine array, was measured in triplicate using an ELISA kit (R&D Systems, Minneapolis, MN, USA).
Statistical analysis
Significance of differences in cell viability, cell growth, intensity of protein spots in 2D-DIGE, and humoral factor secretion was calculated using Student’s t-test.
Results
Expression of all the NMDA receptor subunits in GIMEN cells
We first examined which subunits of NMDA receptors GIMEN cells expressed. As a result, all the subunits of NR1, NR2A-D, and NR3A, B were expressed on GIMEN cells (Figure 1A). Thus, memantine was considered to affect GIMEN cells by binding NMDA receptors.
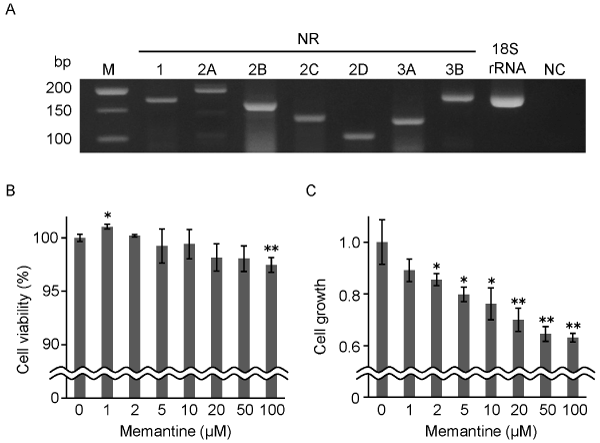
Figure 1. Effects of memantine on the viability and growth of GIMEN cells. A. Expression of NMDA receptor subunits in GIMEN cells was examined by RT-PCR. 18S ribosomal RNA was used as a template as positive control. Distilled water was similarly used as negative control (NC). M, DNA size marker. B. The viability of GIMEN cells cultured for 48 hours in the presence or absence of 1-100 mM memantine. Relative values of the viability to that in the absence of memantine are shown.
C. The growth of GIMEN cells cultured for 48 hours in the presence or absence of 1-100 mM memantine. Relative values of the growth to that in the absence of memantine are shown. All the experiments were performed in triplicates. Error bars, standard deviations. *p<0.05, **p<0.01.
Effects of memantine on the viability and growth of GIMEN cells
To investigate effects of memantine on the viability and growth of GIMEN cells, the cells were treated with 1-100 mM memantine under FBS-free condition. The viability of GIMEN cells was 99.0% or more in the range of 1-10 mM memantine compared that in the absence of memantine (100%) (Figure 1B). The viability was slightly increased by 1 mM memantine (101.0%, p<0.05) and slightly decreased by 100 mM memantine (97.5%, p<0.01). On the other hand, the viable cell numbers in the range of 0-100 mM memantine were 4.22 to 2.66 x 104 cells after the 48 hour treatment, all of which was increased from the seeded cell numbers (2.2 x 104 cells). Interestingly, the growth of GIMEN cells was decreased in a dose-dependent manner (2-100 mM, 85.5-63.0%; 2-10 mM, p<0.05; 20-100 mM, p<0.01) (Figure 1C). From these results, we determined to treat GIMEN cells with 10 mM memantine in the subsequent protein profile analysis. This was because the cell viability was high enough (99.4%, p³0.05), while the cell growth was decreased to less than 80% (76.1%, p<0.05) by that concentration (Figure 1B, C).
Change of the protein profile of GIMEN cells by treatment with memantine
We compared protein profiles between 10 μM memantine-treated and non-treated GIMEN cells by 2D-DIGE to find proteins involved in the suppression of cell growth. As a result, 892 protein spots were detected (Figure 2A, B). 45 out of the 892 protein spots showed different intensity between the memantine-treated and non-treated cells (p<0.05) (Table 1). Among the 45 protein spots, 13 protein spots showed 1.2-fold or higher intensity in the memantine-treated cells compared to the non-treated cells. In contrast, 19 protein spots showed -1.2 (1/1.2)-fold or lower intensity in the memantine-treated cells compared to the non-treated cells. Protein profile of GIMEN cells was changed by the treatment with memantine.
Table 1. Change of the protein profile by addition of memantine
Total number of detected protein spots |
Number of protein spots with different intensity (p<0.05) |
Fold difference, Memantine/Control |
Number of the protein spots |
|
|
x ≥ 1.5 |
5 |
892 |
45 |
1.5 > x ≥ 1.3
1.3 > x ≥ 1.2
1.2 > x > -1.2
-1.2 ≥ x > -1.3
-1.3 ≥ x > -1.5 |
6
2
13
11
8 |
|
|
-1.5 ≥ x |
0 |
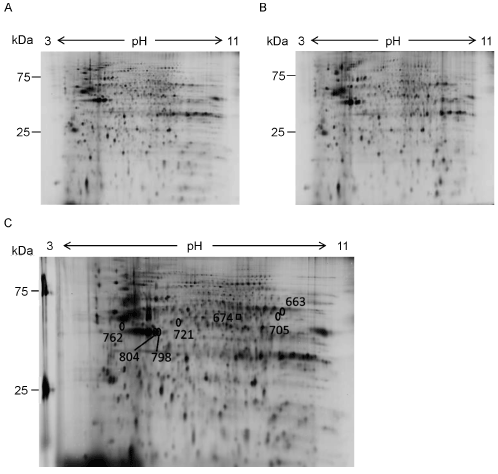
Figure 2. Protein profile of GIMEN cells treated with memantine. A, B. Proteins extracted from 10 mM memantine-treated GIMEN cells (A) and non-treated GIMEN cells (B) were separated by 2D-DIGE. Each analysis was performed in triplicates. Representative 2D-DIGE results are shown. C. Proteins in 7 out of the 32 protein spots, intensity of which were changed ±1.2-fold or more, were identified ( 2). A square and circles show 1 and 6 spots, intensity of which was increased and decreased by the treatment with memantine, respectively.
Identification of the proteins expression of which was changed by memantine
We tried to identify the proteins in the spots, intensity of which was changed by the treatment with memantine. 32 protein spots that showed ±1.2-fold or more intensity change by memantine were subjected to the identification. As a result, 6 proteins were identified from 7 out of the 32 spots (Figure 2C, Table 2). Coronin-1C was identified from the 1 spot, intensity of which was increased 1.44-fold by the treatment with memantine (Figure 2C, Figure 3A, Table 2). In contrast, 5 proteins were identified from the 6 spots, intensity of which was decreased by the treatment with memantine (Figure 2C, Figure 3B-G). They were b-actin (-1.21-fold decreased), g-enolase (-1.21-fold decreased), glutathione synthetase (-1.28-fold decreased), spermatogenesis-associated protein 24 (-1.46-fold decreased), and V-set transmembrane domain-containing protein 2B (-1.46-fold decreased). b-actin was identified from 2 spots (ID 798 and ID 804).
Table 2. Identification of protein spots intensity of which was changed by addition of memantine
Spot |
Fold |
Proteins |
Accession ID |
MW (kDa) |
pI |
Mascot |
Coverage |
Confirmed sequences |
ID |
difference |
|
|
Theoret |
Observed |
Theoret |
Observed |
scores |
(%) |
(Mascot ion scores) |
674 |
1.44 |
Coronin-1C |
COR1C_HUMAN |
53.2 |
53 |
6.65 |
6.7 |
74 |
21 |
224AIFLADGNVFTTGFSR239 (25) |
798 |
-1.21 |
Actin, cytoplasmic 1 |
ACTB_HUMAN |
41.7 |
42 |
5.29 |
5.4 |
195 |
25 |
29AVFPSIVGRPR39 (23) |
|
|
|
|
|
|
|
|
|
|
239SYELPDGQVITIGNER254 (111) |
|
|
|
|
|
|
|
|
|
|
360QEYDESGPSIVHR372 (14) |
804 |
-1.21 |
Actin, cytoplasmic 1 |
ACTB_HUMAN |
41.7 |
42 |
5.29 |
5.35 |
182 |
31 |
29AVFPSIVGRPR39 (6) |
|
|
|
|
|
|
|
|
|
|
239SYELPDGQVITIGNER254 (97) |
|
|
|
|
|
|
|
|
|
|
360QEYDESGPSIVHR372 (28) |
762 |
-1.21 |
Gamma-enolase |
ENOG_HUMAN |
47.2 |
48 |
4.91 |
5.1 |
69 |
16 |
33AAVPSGASTGIYEALELR50 (8) |
|
|
|
|
|
|
|
|
|
|
240IVIGM*DVAASEFYR253 (4) |
|
|
|
|
|
|
|
|
|
|
270YITGDQLGALYQDFVR285 (19) |
721 |
-1.28 |
Glutathione synthetase |
GSHB_HUMAN |
52.4 |
50 |
5.67 |
5.5 |
67 |
15 |
113EGIAQTVFLGLNR125 (7) |
|
|
|
|
|
|
|
|
|
|
142QIEINTISASFGGLASR158 (8) |
|
|
|
|
|
|
|
|
|
|
254LFVDGQEIAVVYFR267 (24) |
663 |
-1.46 |
Spermatogenesis-associated protein 24 |
SPA24_HUMAN |
23.6 |
55 |
7.77 |
8.0 |
59 |
30 |
24DVIESQEELIHQLR37 (16) |
705 |
-1.46 |
V-set and transmembrane domain-containing protein 2B |
VTM2B_HUMAN |
30.3 |
50 |
7.88 |
7.9 |
69 |
35 |
41EGDDIEMPCAFR52 (26) |
32 protein spots, intensity of which changed to ±1.2-fold or more after addition of memantine, were selected for protein identification. Proteins of 7 out of the 32 spots were identified.
*Oxidation of methionine.
MW, molecular weights.
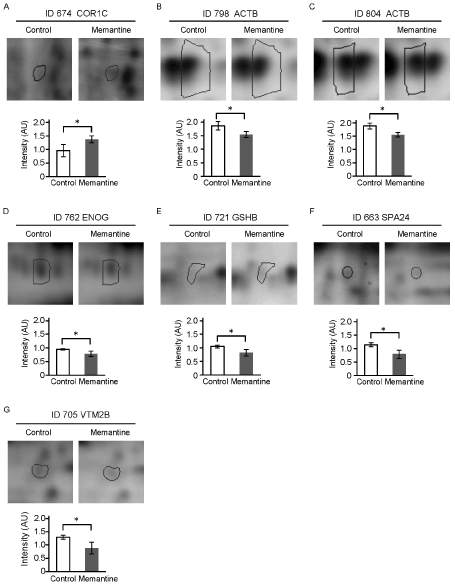
Figure 3. Comparisons of spot intensity of the identified proteins between the memantine-treated and non-treated conditions. Representative results of the identified 7 protein spots and intensity difference of those spots between the memantine-treated and non-treated (control) GIMEN cells are shown.
Error bars, standard deviations. *p<0.05.
Effects of memantine on humoral factor secretion from GIMEN cells
Finally, we compared levels of humoral factor secretion between the memantine-treated and non-treated GIMEN cells. A cytokine array analysis using the culture supernatants showed clear spots of 2 humoral factors, MCP-1 and BDNF (Figure 4A). After the normalization using the positive control spot intensity, intensity of MCP-1 spots tended to be higher in the memantine-treated cells than in the non-treated cells (Figure 4B). We quantified the secreted MCP-1 by ELISA, however, no difference in MCP-1 secretion was found between the memantine-treated and non-treated cells (Figure 4C). Memantine did not affect secretion of the examined 20 humoral factors from GIMEN cells.
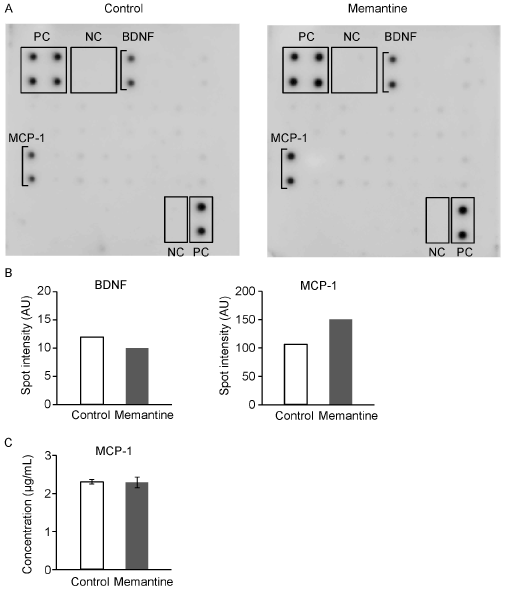
Figure 4. Effects of memantine on humoral factor secretion from GIMEN cells. A. The secretion of humoral factors from GIMEN cells which cultured with medium-alone (left) or 10 mM memantine (right) was assayed using a humoral factor array, in which 20 different anti-humoral factor antibodies were blotted. PC, positive control; NC, negative control. B. The spot intensity of BDNF and MCP-1, which were detected in the humoral factor array results, are shown. C. Concentration of MCP-1 was measured by ELISA. Error bars, standard deviations.
Discussion
In this study, we explored novel actions of memantine by comparing the memantine-treated GIMEN cells to the non-treated cells. GIMEN cells expressed all the 7 NMDA receptor subunits, including NR2B that is related to the glutamatergic excitotoxicity (Figure 1A) [6, 9]. Memantine was considered to affect GIMEN cells through NMDA receptors as well as it affects neuronal cells in the AD brain. The subunit composition of NMDA receptors is known to be plastic [6], therefore, all the 7 subunits may be used to form heterotetrameric complexes of the receptors in various combinations. Protein expression of the NMDA receptor subunits in GIMEN cells should be further confirmed.
The viability of GIMEN cells was not decreased by memantine except the 100 mM treatment (97.3%, p<0.01) (Figure 1B). This result was different from the previous study using another neuroblastoma cell line, SK-N-SH [16]. The viability of SK-N-SH cells was relatively increased by 10 mM and 100 mM memantine [16]. Since SK-N-SH cells do not express functional NMDA receptors [22], memantine may decrease the viability of GIMEN cells through NMDA receptors.
In contrast, the growth of GIMEN cells was apparently decreased in a dose-dependent manner in the range of 2-100 mM memantine (2-10 mM, 85.5-76.1%, p<0.05; 20-100 mM, 70.0-63.0%, p<0.01) (Figure 1C). Thus, memantine was considered to suppress the growth of GIMEN neuroblastoma cells by its pharmacological effect. In several neuroblastoma cell lines, mRNA expression of NR1 subunit of NMDA receptors was shown, however, the expression of NR2A-D was not shown or was controversial among the studies [22-24]. In contrast, protein expression of NMDA receptors and/or their involvement in cell proliferation have been reported in other types of tumor cells such as glioma, prostatic cancer, breast cancer, colon cancer, and small cell lung cancer [25-27]. In such tumor cells, 20-900 mM memantine reduced the cell growth probably by limiting signals from NMDA receptors. In our results, relatively low doses of 2-50 mM memantine showed suppression of the cell growth without affecting the cell viability (Figure 1B, C). Interestingly, 1 mM memantine that is within therapeutic serum concentrations for AD [28] also tended to suppress the cell growth (89.1%, p³0.05) (Figure 1C). Memantine may have an effect on the inhibition of neuroblastoma cell growth. On the other hand, the therapeutic concentration of memantine, 1mM, may suppress growth of normal neural cells in adult brains, i.e., the growth of adult neural stem/progenitor cells. The suppressive effect on GIMEN cell growth should be validated using other neuroblastoma cell lines expressing NMDA receptors and also using normal neural cells.
Under therapeutic condition, serum concentrations of memantine in man with daily maintenance doses of 20 mg range from 0.5-1.0 mM, while the concentrations in cerebrospinal fluid (man) and brain microdialysates (rat) are 20-50% lower due to albumin binding in serum [28]. However, the brain homogenates in both man and rodents contain much higher concentrations of memantine (10-30 fold) probably due to lysosomal accumulation [28]. Recently, it was found that macroautophagy occurs in neurons of an AD mouse model to increase Aβ incorporation into secondary lysosomes [29]. A part of such neurons undergo atypical cell death to release undegraded Aβ to extracellular space [29]. In such condition, neurons can be exposed to higher concentrations of memantine similarly released from lysosomes of the dead cells. Taken together the above-mentioned reasons with the results of cell viability and growth, we determined the concentration of memantine to be 10 mM for the analysis of protein expression. Among 892 protein spots detected in the 2D-DIGE results, intensity of 32 spots was changed ±1.2-fold or more between the memantine-treated and non-treated GIMEN cells (p<0.05) (Table 1). The number of the protein spots, intensity of which was changed, was comparable with those in the other proteomic studies that similarly examined actions of drugs or cytokines [30, 31].
We identified proteins in 7 out of the 32 spots and found the decreased expression of b-actin, g-enolase, and glutathione synthetase by the treatment with memantine (Table 2, Figure 3B-E). b-actin, a cytoskeletal protein, involves in cytokinesis as well as in cell migration and cell morphogenesis [32]. The decreased expression of b-actin is considered to be associated with the suppression of GIMEN cell growth. In contrast, expression of coronin-1C that contains multiple actin-binding sites [33] was increased by the treatment with memantine (Table 2, Figure 3A). Since coronin-1C has been suggested to act as an integrator of actin filaments [34], coronin-1C may be increased to recover the down-regulated functions of b-actin. g-enolase, predominantly expressed in neurons and cells of the neuroendocrine system, is an enzyme of the glycolytic pathway which catalyzes the dehydration of 2-phospho-D-glycerate to phosphoenol-pyruvate [35]. g-enolase promotes survival of neuronal cells by regulating neuronal growth factor receptor-dependent signaling pathway, which results in extensive actin cytoskeleton remodeling [35]. Interestingly, p19ras was suggested to suppress proliferation of cancer cells through the interaction with g-enolase [36]. The decrease of g-enolase may be also involved in the suppression of GIMEN cell growth, relating to the decrease of b-actin.
Glutathione is a tripeptide formed by glutamate, cysteine, and glycine, which plays an important role in a multitude of cellular processes, including cell proliferation and apoptosis [37]. Glutathione synthetase is involved in the subpathway of glutathione biosynthesis, in which glutathione is synthesized from g-glutamylcysteine and glycine. Glutathione is required for cell proliferation and is essential for the cells to enter the S phase [38, 39]. The level of glutathione is up-regulated by glutathione synthetase and is correlated with cell growth rate [39]. The decrease of glutathione synthetase by the treatment with memantine would reduce the glutathione level in GIMEN cells, which may further lead to the suppression of cell growth.
Effects of memantine on molecular profiles of neuronal cells were previously studied by analyses of mRNA expression of rat cortex using cDNA microarray [18] and protein expression and modification of mouse hippocampus and cortex using reverse phase protein arrays [19]. However, none of molecules found in these studies was detected in our study. This may be due to the difference of materials (human neuroblastoma vs mouse or rat brain tissue) and methods (2D-DIGE and mass spectrometry vs cDNA or protein arrays). The effects of memantine on protein profiles found in this study should be validated using other neuroblastoma cells and normal neuronal cells in near future.
In conclusion, we found that memantine suppressed the growth of GIMEN cells and affected the protein profile. Memantine may have an effect to inhibit growth of neuroblastoma cells, whereas it may suppress growth of normal neural cells even at the therapeutic concentrations. The suppression of cell growth may be caused by the decreased expression of b-actin, g-enolase, and glutathione synthase due to the loss of their functions associated with cell proliferation. Further investigations are needed to validate the above results and to elucidate mechanisms of the newly found actions of memantine in this study.
Acknowledgement
The authors are indebted to Ms. Kumi Aso for her technical assistance. The authors are also grateful to Daiichi Sankyo for the support for this study.
Conflict of interest
None.
References
- Scheltens P, Blennow K, Breteler MM, de Strooper B, Frisoni GB, et al. (2016) Alzheimer's disease. Lancet 388: 505-517. [Crossref]
- Magi S, Castaldo P, Macrì ML, Maiolino M, Matteucci A, et al. (2016) Intracellular Calcium Dysregulation: Implications for Alzheimer's Disease. Biomed Res Int 2016: 6701324. [Crossref]
- Hung SY, Fu WM (2017) Drug candidates in clinical trials for Alzheimer's disease. J Biomed Sci 24: 47. [Crossref]
- Wang T (2017) New Drug Research and Development for Alzheimer's Pathology: Present and Prospect. Shanghai Arch Psychiatry 29: 237-239. [Crossref]
- Rosini M, Simoni E, Minarini A, Melchiorre C (2014) Multi-target design strategies in the context of Alzheimer's disease: acetylcholinesterase inhibition and NMDA receptor antagonism as the driving forces. Neurochem Res 39: 1914-1923. [Crossref]
- Paoletti P, Bellone C, Zhou Q (2013) NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci 14: 383-400. [Crossref]
- Albensi BC (2007) The NMDA receptor/ion channel complex: a drug target for modulating synaptic plasticity and excitotoxicity. Curr Pharm Des 13: 3185-3194. [Crossref]
- Rezvani AH (2006) Involvement of the NMDA System in Learning and Memory. In: Levin ED, Buccafusco JJ, editors. Animal Models of Cognitive Impairment. Boca Raton (FL): CRC Press/Taylor & Francis. Chapter 4.
- Rudy CC, Hunsberger HC, Weitzner DS, Reed MN (2015) The role of the tripartite glutamatergic synapse in the pathophysiology of Alzheimer's disease. Aging Dis 6: 131-148. [Crossref]
- Lewerenz J, Maher P (2015) Chronic Glutamate Toxicity in Neurodegenerative Diseases-What is the Evidence? Front Neurosci 9: 469. [Crossref]
- Bello O, Blair K, Chapleau C, Larimore JL (2013) Is memantine a potential therapeutic for Rett syndrome? Front Neurosci 7: 245. [Crossref]
- Zdanys K, Tampi RR (2008) A systematic review of off-label uses of memantine for psychiatric disorders. Prog Neuropsychopharmacol Biol Psychiatry 32: 1362-1374. [Crossref]
- Rogawski MA, Wenk GL (2003) The neuropharmacological basis for the use of memantine in the treatment of Alzheimer's disease. CNS Drug Rev 9: 275-308. [Crossref]
- Lipton SA (2004) Paradigm shift in NMDA receptor antagonist drug development: molecular mechanism of uncompetitive inhibition by memantine in the treatment of Alzheimer's disease and other neurologic disorders. J Alzheimers Dis 6: S61-74. [Crossref]
- Ray B, Banerjee PK, Greig NH, Lahiri DK (2010) Memantine treatment decreases levels of secreted Alzheimer's amyloid precursor protein (APP) and amyloid beta (A beta) peptide in the human neuroblastoma cells. Neurosci Lett 470: 1-5. [Crossref]
- Alley GM, Bailey JA, Chen D, Ray B, Puli LK, et al. (2010) Memantine lowers amyloid-ß peptide levels in neuronal cultures and in APP/PS1 transgenic mice. J Neurosci Res 88: 143-154. [Crossref]
- Li L, Sengupta A, Haque N, Grundke-Iqbal I, Iqbal K (2004) Memantine inhibits and reverses the Alzheimer type abnormal hyperphosphorylation of tau and associated neurodegeneration. FEBS Lett 566: 261-269. [Crossref]
- Marvanová M, Lakso M, Wong G (2004) Identification of genes regulated by memantine and MK-801 in adult rat brain by cDNA microarray analysis. Neuropsychopharmacology 29: 1070-1079. [Crossref]
- Ahmed MM, Dhanasekaran AR, Block A, Tong S, Costa ACS, et al. (2014) Protein profiles associated with context fear conditioning and their modulation by memantine. Mol Cell Proteomics 13: 919-937. [Crossref]
- Anaparti V, Ilarraza R, Orihara K, Stelmack GL, Ojo OO, et al. (2015) NMDA receptors mediate contractile responses in human airway smooth muscle cells. Am J Physiol Lung Cell Mol Physiol 308: L1253-L1264.
- Katano M, Kurokawa MS, Matsuo K, Masuko K, Suematsu N, et al. (2017) Phosphoproteome analysis of synoviocytes from patients with rheumatoid arthritis. Int J Rheum Dis 20:708-721.[Crossref]
- Pizzi M, Boroni F, Bianchetti A, Moraitis C, Sarnico I, et al. (2002) Expression of functional NR1/NR2B-type NMDA receptors in neuronally differentiated SK-N-SH human cell line. Eur J Neurosci 16: 2342-2350. [Crossref]
- Yoshioka A, Ikegaki N, Williams M, Pleasure D (1996) Expression of N-methyl-D-aspartate (NMDA) and non-NMDA glutamate receptor genes in neuroblastoma, medulloblastoma, and other cell lines. J Neurosci Res 46: 164-178. [Crossref]
- North WG, Fay MJ, Du J, Cleary M, Gallagher JD, et al. (1997) Presence of functional NMDA receptors in a human neuroblastoma cell line. Mol Chem Neuropathol 30: 77-94. [Crossref]
- Takano T, Lin JH, Arcuino G, Gao Q, Yang J, et al. (2001) Glutamate release promotes growth of malignant gliomas. Nat Med 7:1010-1015. [Crossref]
- Abdul M, Hoosein N (2005) N-methyl-D-aspartate receptor in human prostate cancer. J Membr Biol 205: 125-128. [Crossref]
- North WG, Gao G, Jensen A, Memoli VA, Du J (2010) NMDA receptors are expressed by small-cell lung cancer and are potential targets for effective treatment. Clin Pharmacol 2: 31-40. [Crossref]
- Parsons CG, Danysz W, Quack G (1999) Memantine is a clinically well tolerated N-methyl-D-aspartate (NMDA) receptor antagonist--a review of preclinical data. Neuropharmacology 38: 735-767. [Crossref]
- Chen X, Kondo K, Motoki K, Homma H, Okazawa H (2015) Fasting activates macroautophagy in neurons of Alzheimer's disease mouse model but is insufficient to degrade amyloid-beta. Sci Rep 5: 12115.
- Endo W, Arito M, Sato T, Kurokawa MS, Omoteyama K, et al. (2014) Effects of sulfasalazine and tofacitinib on the protein profile of articular chondrocytes. Mod Rheumatol 24: 844-850. [Crossref]
- Tsuno H, Suematsu N, Sato T, Arito M, Matsui T, et al. (2016) Effects of methotrexate and salazosulfapyridine on protein profiles of exosomes derived from a human synovial sarcoma cell line of SW982. Proteomics Clin Appl 10: 164-171. [Crossref]
- Pollard TD, Cooper JA (2009) Actin, a central player in cell shape and movement. Science 326: 1208-1212. [Crossref]
- Tilley FC, Williamson RC, Race PR, Rendall TC, Bass MD (2015) Integration of the Rac1- and actin-binding properties of Coronin-1C. Small GTPases 6: 36-42. [Crossref]
- Behrens J, Solga R, Ziemann A, Rastetter RH, Berwanger C, et al. (2016) Coronin 1C-free primary mouse fibroblasts exhibit robust rearrangements in the orientation of actin filaments, microtubules, and intermediate filaments. Eur J Cell Biol 95: 239-251. [Crossref]
- Vizin T, Kos J (2015) Gamma-enolase: a well-known tumor marker, with a less-known role in cancer. Radiol Oncol 49: 217-226. [Crossref]
- Jang S, Kim J, Kim C, Kim D, Rhee S, et al. (2010) p19ras represses proliferation of non-small cell lung cancer possibly through interaction with neuron-specific enolase (NSE). Cancer Lett 289: 91-98. [Crossref]
- Traverso N, Ricciarelli R, Nitti M, Marengo B, Furfaro AL, et al. (2013) Role of glutathione in cancer progression and chemoresistance. Oxid Med Cell Longev 2013: 972913. [Crossref]
- Messina JP, Lawrence DA (1989) Cell cycle progression of glutathione-depleted human peripheral blood mononuclear cells is inhibited at S phase. J Immunol 143: 1974-1981. [Crossref]
- Huang Z, Chen C, Zeng Z, Yang H, Oh J, et al. (2001) Mechanism and significance of increased glutathione level in human hepatocellular carcinoma and liver regeneration. FASEB J 15: 19-21. [Crossref]




