Abstract
The development of antibiotic resistance in livestock species is discussed as major threat to modern medicine. One potential strategy to battle resistance in animal production might be the application of feed additives with antimicrobial properties, such as organic acids. The aim of this study was to evaluate the impact of an acid-based feed additive on antimicrobial resistance traits of faecal microbiota in weaning piglets and to compare the results to antibiotic intervention with oxytetracycline. Therefore, 24 pigs were allocated to three treatment groups, fed a basal diet and a basal diet supplemented with either the antibiotic or the feed additive. Quantitative PCR was used to monitor the prevalence and quantity of frequently associated genes encoding resistance to tetracycline, streptomycin, and sulfonamide. The number of positive tested samples for tetB and sul1 decreased over time, with differences due to treatment. For all analysed genes, a decrease in log10 mean antibiotic resistance gene copy numbers was observed in all groups on day 10 of the experiment. The mean tetA and tetB gene levels were significantly higher in the oxytetracycline group than in the other groups indicating selection for these antibiotic resistance genes. No effects of the applied feed additive on the quantity of selected antibiotic resistance genes in faeces were detected. In contrast, antibiotic treatment clearly effected antibiotic resistance gene level and prevalence. Results implicate that, in the long run, novel feed additives have the potential to minimise the role of livestock as a reservoir for antimicrobial resistance.
Key words
pigs, antibiotic resistance gene, novel feed additive, qPCR, total bacterial community DNA
Introduction
In the last decades, livestock production has intensified dramatically to meet the rising demand of animal derived foods. In the light of economic production, the use of antibiotics has become an integrated part of the production system. Following the ban of antibiotic growth promoters in the European Union, due to the quick rise of antibiotic resistances, antibiotics are specifically used as a metaphylactic or therapeutic treatment. Strict rules exist how to apply antibiotics in animal production [1]. However, each antibiotic usage might contribute to the risk of antibiotic resistance transfer and dissemination [2].
In pig farming, antibiotic treatment is often applied in young piglets [3] because mortality is high in critical periods of growth, such as the first week of life and immediately after weaning. The post-weaning stress and the related immunocompromised state promote the development of bacterial infections such as E. coli associated diarrhea [4,5]. Chlortetracycline and oxytetracycline are among the most commonly used antibiotics in pig production in the European Union and the United States [6,7]. Therefore, it is not surprising that tetracycline resistance represents the most commonly detected resistance among bacteria. Resistance occurs mainly due to acquisition of tet genes that are involved in the active efflux of the drug or by protecting the ribosome from its action (Roberts, 2011). One main concern is the co-selection of other important resistances. Association of resistance genes with mobile genetic elements allows the accumulation of a large number of different resistance genes or other types of genes, which might be kept in the bacterial communities even without selective pressure. Among Gram-negative tet genes, the widespread tetA and tetB genes have been shown to be linked to other resistance genes. They are frequently associated with genes conferring resistance to streptomycin (strA, strB) and/or sulfamethoxazole (sul1, sul2) [8].
Due to the risk of gene transfer, antibiotic resistance in livestock species is discussed as major risk to public health. Besides improvement of biosecurity and instalment of vaccination programs, the use of novel feed additives could help to reduce antibiotics in livestock production and at the same time maintain animal health and productivity [9]. Organic acids were reported to stimulate the digestive system of pigs and to improve pig performance because of their antimicrobial activity [10-12]. Several studies show that certain organic acids can inhibit growth of multi-resistant bacteria and that they can act against bacterial biofilms, which cannot be achieved by antibiotics [13,14]. The impact of acid-based feed supplements on antimicrobial resistance traits has not been investigated in pigs so far. The aim of our study was to compare the effect of additionally feeding oxytetracycline and an acid-based feed additive on the development of resistance gene levels in weaned piglets. Quantitative PCR (qPCR) was used to assess the prevalence and quantity of frequently linked genes conferring resistance to tetracyclines, aminoglycosides and sulfonamides in faeces samples.
Materials and methods
Animals and housing
The experiment was conducted in the experimental centre of farm animals at the Slovak University of Agriculture in Nitra. All procedures related to the experiment were carried out following the European Guidelines for the Care and Use of Animals for Research Purpose [15]. The animal experiment was approved by the State Veterinary and Food Administration of the Slovak Republic (project number 1485/15-221).
After weaning, 24 Large White piglets (mixed sex, four weeks of age) were randomly distributed to three treatment groups (n=8), taking into account the body weight, sex, and mother sow. Animals were housed pairwise in pens on concrete floor under controlled environmental conditions and had free access to water during the whole duration of the trial. Piglets as well as their dams had no history of antibiotic usage.
Dietary treatment and sampling
Pigs received one of three different dietary treatments in dry form ad libitum: a basal diet serving as control group (CON; without supplements), a basal diet supplemented with the antibiotic oxytetracycline (OTC; “Chevita”, Chevita GmbH, Germany) or a basal diet supplemented with a commercial feed additive consisting of a blend of propionic, formic and acetic acid, cinnamaldehyde and a permeabilizing complex (FA; BIOTRONIC TOP3, BIOMIN Holding GmbH, Austria, 2 kg/t). The antibiotic was administered directly after weaning for 10 days in a dose recommended by the manufacturer (40 mg oxytetracycline hydrochloride/kg body weight/day). Individual faecal samples were collected from the rectum at day 0, 10 and 28, respectively. Samples were placed into individual tubes, snap frozen on dry ice and stored at -40 °C until further usage.
Bacterial community DNA extraction of faeces
Total bacterial community DNA was extracted from 200 mg of frozen faecal samples using the QIAamp DNA Stool Mini Kit (QIAgen, USA). After addition of 1.4 mL buffer ASL, a mechanical lysis step using the homogenizer Precellys 24 (Bertin, USA) at 6800 g for 15 s was included in the protocol before heating the suspension for 5 min at 95 °C. Further steps were performed according to the manufacturer’s instructions.
Quantification of the total bacterial load and antibiotic resistance genes
The above-prepared DNA was quantified and adjusted to 4 ng/µl for subsequent qPCR analysis of antibiotic resistance genes encoding resistance to tetracycline (tetA, tetB), streptomycin (strA, strB) and sulfonamides (sul1, sul2). To measure the total bacterial load, the 16S ribosomal RNA (rRNA) gene was quantified.
All qPCR assays were performed on a Mastercycler Realplex 2S (Eppendorf, Germany) using SYBR Green detection chemistry. Details on primers and thermal cycling conditions are given in (Table 1). Each reaction was carried out in a total volume of 15 μL, containing 2 μL of DNA template, 1.5 µL of each primer (1.5 pmol/µL), 7.5 µL of the 2x Kapa SYBR Fast master mix (Biosystems, USA) and 2.5 µL of nuclease-free water. The thermal profile consisted of initial activation at 95 °C for 3 min followed by 40 cycles of denaturation at 95°C for 15s, annealing at primer specific temperature for 20 s and extension at 72 °C for 20 s.
Standard curves were generated with known quantities of target genes cloned into pGEM-T easy plasmids (Promega Corporation, USA). The gene copy number was calculated as described previously [16] and expressed as gene copies per gram faeces.
All samples, standards and controls (non-template control and positive control) were run in triplicates.
Standard curves were further used to define the limit of quantification (LOQ) and limit of detection (LOD) for each individual qPCR assay. The LOQ was defined as the most diluted DNA standard before cycle threshold (Ct) values deviated from the linear range of the standard curve. The LOD was set as the lowest concentration in the standard serial dilution where at least one of the triplicates was positive or before the Ct deviated from the average Ct of a false positive.
Data Management and Statistical Analysis
Faecal samples with resistance gene quantities below the specified LOQ and above the LOD were assigned half the LOQ. Sample concentrations below the LOD were reported as not detected and not included in analysis.
All statistical tests were performed using the software IBM SPSS statistics, Version 22.0 (IBM Corp., USA). The individual pig was used as experimental unit to test the influence of dietary treatment and the sampling time on antibiotic resistance gene quantification. Gene quantities were log10 transformed to achieve normal distribution. Levene's test was performed to verify homogeneity of variances. A one-way analysis of variance (ANOVA) was used to estimate overall differences between the group means, which were further compared using Bonferroni test. In the case of variance inhomogeneity, Welch’s robust test was used to compare group means followed by Tamhane post-hoc test. Differences at P<0.05 were considered significant.
Results
Quantification of the 16S rRNA gene as measure of the total bacterial load revealed a stable microbial population in the piglets of the trial (Table 2). At the start of the experiment mean log 16S rRNA gene copy numbers in faecal samples ranged from 10.83 to 10.93 in the individual treatment groups followed by a minor decrease on day 10 (ranging from 10.77 to 10.80) and increase on day 28 (10.94 to 11.00). A significant difference was found in the CON group between day 10 and day 28 (P=0.012). Since trends described for the gene copy estimates did not change after normalisation with the 16S rRNA gene, only the absolute quantification was taken for further analysis.
As not all targeted antibiotic resistance genes were detected in each sample over the entire trial period, prevalence of antibiotic resistance genes was also taken into account (Table 3). Among the analysed resistance genes, tetA, sul2, strA and strB were detected in faecal samples from all animals, except two where strA and one where strB could not be detected. TetB and sul1 genes were found in all samples before treatment. The number of tetB and sul1- positive samples decreased during the experiment, revealing differences based on treatment. Compared to the CON and the FA group, the number of tetB- positive samples was higher in OTC-fed piglets on day 10 and 28. The sul1 gene was detected in all samples of the treatment groups on day 0 and day 10. At the last time point, it was found just in two samples of the FA group, whereas six and seven samples of the CON and OTC group were positive, for sul1.
Figure 1 displays the mean log10 copy number of tetA, tetB, strA, strB, sul1 and sul2 genes over time (days 0, 10, 28) in response to treatments. At start of the experiment, the mean log10 copies of individual antibiotic resistance genes were in a similar range for the CON, OTC and FA group. On day 10, a decrease in all copy numbers was found, respectively, for all groups. ANOVA analysis revealed a statistically significant time-dependent effect. The levels of tetA and tetB genes turned out to be significantly reduced in faeces of the CON (tetA P=0.033, tetB P=0.001) and the FA group piglets (tetA P=0.007; tetB P=0.011). For strA and strB a significant time-dependent decrease was found in faeces of the CON (strA P=0.02, strB P=0.001) and the OTC group piglets (strA P<0.001, strB P<0.001). Over time, there was a significant reduction of sul1 only in samples from the FA group (P=0.046), whereas sul2 gene copies were significantly decreased in CON and OTC samples (P=0.001, P<0.001).
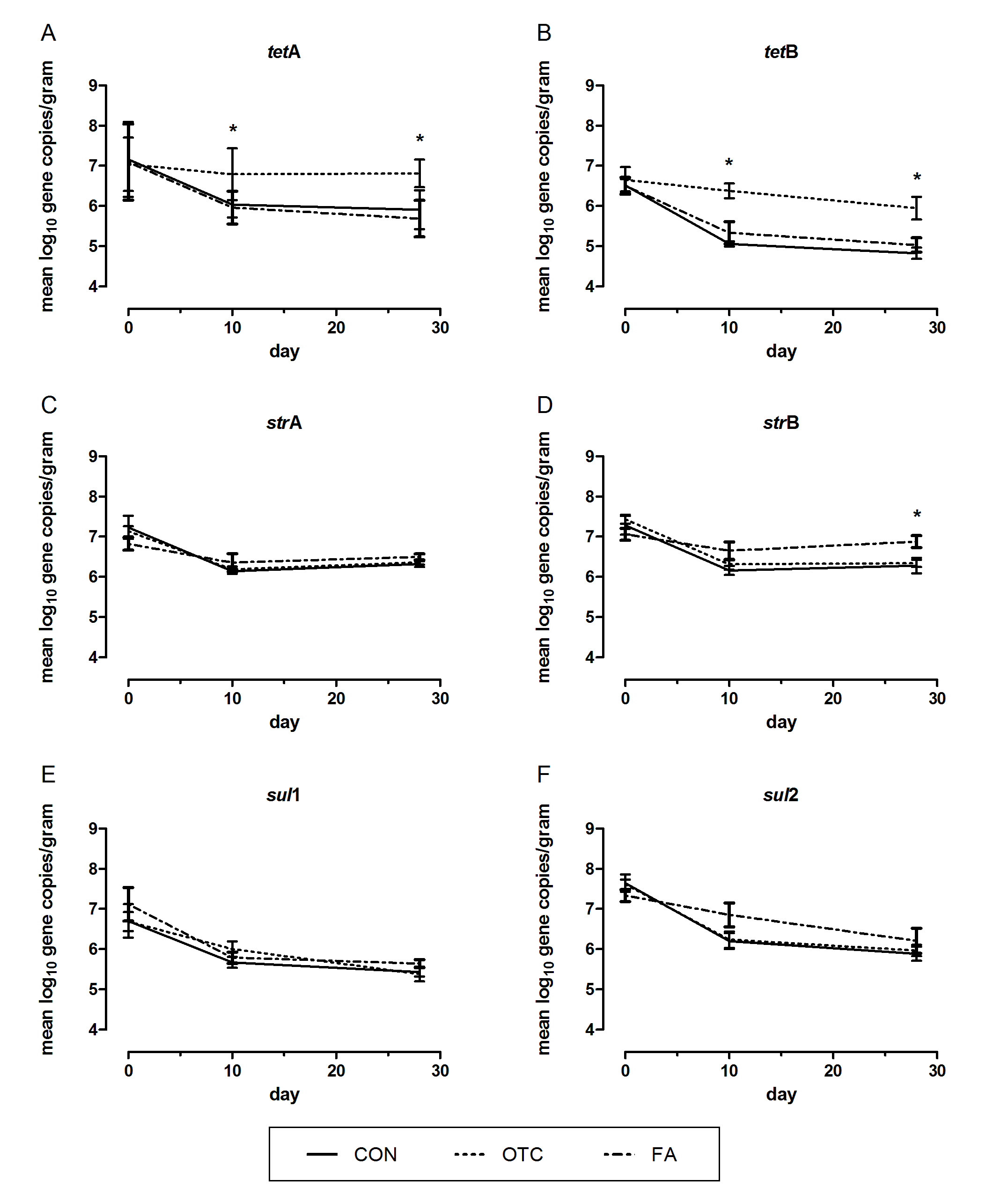
Figure 1. Non-standardized log10 copies of antibiotic resistance genes resistance genes (A) tetA, (B) tetB, (C) strA, (D) strB, (E) sul1 and (D) sul2, per gram of fecal sample from weaning piglets receiving a basal diet (CON) and a basal diet supplemented with oxytetracycline (OTC) or an acid-based feed additive (FA). Mean values with standard deviation are shown. *Superscripts indicate treatment specific differences (P<0.05).
Looking at the effect of the treatments, the mean tetA and tetB gene quantities turned out to be significantly higher in OTC group samples compared to those of the other groups on both day 10 (towards CON P=0.015, P<0.001; towards FA P=0.007, P=0.035) and day 28 (CON P=0.001, P=0.025 and FA P<0.001). On day 28, significant differences were obtained only between the OTC and CON group samples (P=0.025). Streptomycin resistance gene quantities were also affected, with higher strB levels in piglets of the FA compared to the OTC group on day 28 (P=0.034).
Discussion
Recent publications highlight the importance to evaluate positive and negative impacts of various alternatives to antibiotics on the antimicrobial resistance in animal production [17-19]. Assessment of antibiotic resistance in various environments has long relied on culture-based approaches focusing on specific cultivable species. Because a significant proportion (60-80%) of the faecal microbiota is not cultivable [20,21], we used PCR amplification to investigate the prevalence and persistence of antimicrobial resistance genes. This strategy might provide more accurate information on the actual magnitude of antibiotic resistance in the microbial population. The genetic determinants of resistance that were targeted within this experiment consisted of those being exclusively or at least commonly reported in Gram-negative enteric bacteria. The most common resistance mechanism in Gram-negative bacteria is the energy-dependent efflux pump system, which is encoded by diverse tet genes, with tetA and tetB being the most frequently described [22]. These genes are predominantly found in tetracycline resistant Enterobacteriaceae, including members of non-pathogenic microbiota as well as pathogens. They are usually located on large conjugative plasmids, which also carry other antibiotic determinants [23]. Various combinations with antibiotic resistance genes for streptomycin (strA, strB) and sulfonamides (sul1, sul2) are reported and they often cluster on plasmids, integrons or transposons [24,25].
In all faecal samples, 16S rRNA gene concentrations remained stable over the entire experiment, irrespective of pig age and dietary treatment. The quantification of the 16S rRNA gene has been used to normalise the abundance of resistance genes to the bacterial population and to account for variations in DNA extraction efficiency [26]. Because in this experiment no differences based on normalisation were obtained, data of the absolute quantification are reported. In the beginning of the study, all faecal samples were positive for the selected resistance genes, although the piglets as well as their dams were never exposed to any antibiotics. A similar situation has been found in previous studies [27,28], which also reported the presence of background resistances in the absence of any antibiotic selective pressure. In our study, resistance gene copy numbers were higher in the pigs as in the sows (unpublished results) but decreased with time which might reflect age related differences in the microbiota. Faecal bacterial communities are in general dominated by two major phyla, Firmicutes and Bacteroidetes. As pigs age, the proportion of Gram-negative bacteria is decreasing and a shift towards a Gram-positive microbiota is reported [29]. Thus, it might be speculated that the declining trend in the level of resistance genes might be a result of the expansion of Gram-positive bacteria in the underlying bacterial population. The levels of tet genes were not reduced in faeces of piglets additionally administered oxytetracycline, which was indicated by stable tetA and tetB log10 gene copy numbers at both time points (d10, d28), compared to declining ones in non-antibiotic treatments. Previous studies reported elevated levels of resistant bacteria or resistance genes under selective pressure, revealing a clear association between antibiotic consumption and antibiotic resistance [30-32]. Interestingly, shedding of tetraycline resistant bacteria in faecal samples continued after withdrawal of the antibiotic, indicating a long-lasting effect of antibiotic supplementation.
Also, when the occurrence of antibiotic resistance genes in individual samples instead of gene quantity was analysed, a clear effect of oxytetracycline supplementation on faecal prevalence of tetB was detected, at both time points (d10, d28). Although tetB is known to have the widest host range in Gram-negative bacteria among the different tet genes [23], having no selective pressure, a clear reduction of positive samples was observed for CON and FA group piglets. In contrast, tetA remained stable showing presence in 100% of the samples over the entire trial period. In enterobacterial isolates, a strong negative correlation of tetA and tetB was observed for the distribution of these genes, indicating a potential plasmid incompatibility [33]. Although both genes encode for an efflux pump in the lipid bilayer of the bacteria, tetB might be associated with higher levels of resistance, presence of multiple resistance genes and intensive farming conditions [34]. Lee and coworkers [35] even report higher minimal inhibitory concentrations towards tetracycline associated with tetB. Yet, in the present study, the OTC feed supplementation seems to have favoured tetA over tetB, which was also reported in a previous study published by Agga et al. [27].
No treatment specific differences were observed for the gene quantity of sul1. However, a decreased faecal prevalence in the FA, and to a lesser extent in the CON group, might indicate a correlation of tetB and sul1 for a subset of the resistant bacterial population. Genetic linkage of selected antibiotic resistance genes is common. For E. coli, respectively, association of tetA and sul1 is more frequently reported [8].
Interestingly, strB was the only gene, with increased levels in the FA group, only on day 28, when comparing its log10 gene copy numbers to those of the OTC group piglets. Both streptomycin resistance determinants, strA and strB, are widely distributed in Gram-negative bacteria and were found to be linked together on the same transcriptional unit [36]. Yet, quantity of strA was not affected by the acid-based feed supplement. Applied qPCR analyses are not suitable to make conclusions on the bacterial level, though correlation of individual assays might indicate specific patterns of multi- and co-resistance, but do not constitute a standalone approach in this regard. However, as a total community-based approach qPCR gives an overall picture of the resistance level and constitutes a reliable tool to easily check for antibiotic resistance genes in environmental samples and might be used for evaluating the impact of growth promoters in livestock farming.
Conclusion
Clear effects of the antibiotic treatment on the resistance gene prevalence and quantity were observed. An obvious reduction in the quantity of selected antibiotic resistance genes by the acid-based feed additive was not detected in the present study. Still, antimicrobial activities of organic acids are well known, and therefore, their potential to reduce bacterial antimicrobial resistance should be further investigated. Upcoming experiments should more thoroughly study the role of the gut microbiota and/or mobile genetic elements such as plasmids, transposons and integrons in the dissemination of resistance genes in pigs over time and the effect of feed supplements on this dissemination.
Conflict of interests
BIOMIN Holding GmbH provided support in the form of salaries for authors GW, VK and VN, but did not have any additional role in the experimental design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors declare no conflict of interest.
Acknowledgments
The authors appreciate the excellent animal care skills of the staff at the Department of Animal Nutrition at Slovak University of Agriculture in Nitra during conduction of the feeding trial. Finally, the authors wish to thank Barbara Doupovec for assistance in statistical analysis and Barbara Weber for critical reviewing the manuscript.
References
- European Commission (2015) Guidelines for the prudent use of antimicrobials in veterinary medicine (C 299). Official Journal of the European Union: 1-20.
- Aarestrup FM, Oliver Duran C, and Burch DGS (2008) Antimicrobial resistance in swine production. Anim Health Res Rev 9: 135-148. [Crossref]
- Sjölund M, Postma M, Collineau L, Lösken S, Backhans A, et al. (2016) Quantitative and qualitative antimicrobial usage patterns in farrow-to-finish pig herds in Belgium, France, Germany and Sweden. Prev Vet Med 130: 41-50. [Crossref]
- Heo JM, et al. (2013) Gastrointestinal health and function in weaned pigs: a review of feeding strategies to control post-weaning diarrhoea without using in-feed antimicrobial compounds. J Anim Physiol Anim Nutr (Berl) 97: 207-237. [Crossref]
- Melin L, Mattsson S, Katouli M, Wallgren P (2004) Development of post-weaning diarrhoea in piglets. Relation to presence of Escherichia coli strains and rotavirus. J Vet Med B Infect Dis Vet Public Health 51: 12-22. [Crossref]
- Apley MD, Bush EJ, Morrison RB, Singer RS, Snelson H (2012) Use estimates of in-feed antimicrobials in swine production in the United States. Foodborne Pathog Dis 9: 272-279. [Crossref]
- European Medicines Agency (2016) Sales of veterinary antimicrobial agents in 26 EU/EEA countries in 2014. In: European Surveillance of Veterinary Antimicrobial Consumption.
- Boerlin P, Travis R, Gyles CL, Reid-Smith R, Janecko N, et al. (2005) Antimicrobial resistance and virulence genes of Escherichia coli isolates from swine in Ontario. Appl Environ Microbiol 71: 6753-6761. [Crossref]
- de Lange CFM, Pluske J, Gong J, and Nyachoti CM (2010) Strategic use of feed ingredients and feed additives to stimulate gut health and development in young pigs. Livestock Science 134: 124-134.
- Papatsiros VG and Billinis C (2012) The Prophylactic Use of Acidifiers as Antibacterial Agents in Swine. In: V Bobbarala (ed.), Antimicrobial Agents. Rijeka (Croatia): InTech.
- Taube VA, et al. (2009) Effects of dietary additives (potassium diformate/organic acids) as well as influences of grinding intensity (coarse/fine) of diets for weaned piglets experimentally infected with Salmonella Derby or Escherichia coli. J Anim Physiol Anim Nutr (Berl) 93: 350-358. [Crossref]
- Riemensperger AV, Bachinger D, Schaumberger S, Urbaityte R, and Pasteiner S (2012) The effect of an organic acid blend, cinnamaldehyde and a permeabilising substance on the inhibition of bacterial growth in vitroand growth performance of weaning pigs. Veterinarija ir zootechnika 60: 59-66.
- Goualié BG, et al. (2014) Occurrence of multidrug resistance in Campylobacter from Ivorian poultry and analysis of bacterial response to acid shock. Food Science and Biotechnology 23: 1185-1191.
- Akbas MY, Cag S (2016) Use of organic acids for prevention and removal of Bacillus subtilis biofilms on food contact surfaces. Food Sci Technol Int 22: 587-597. [Crossref]
- European Commission (2010) Directive 2010/63/EU on the protection of animals used for scientific purposes (L 276). Official Journal of the European Union: 1-47.
- Pei R, Kim SC, Carlson KH, and Pruden A (2006) Effect of River Landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res 40: 2427-2435.
- Aperce CC, Amachawadi R, Van Bibber-Krueger CL, Nagaraja TG, Scott HM, et al. (2016) Effects of Menthol Supplementation in Feedlot Cattle Diets on the Fecal Prevalence of Antimicrobial-Resistant Escherichia coli. PLoS One 11: e0168983. [Crossref]
- Jahanbakhsh S, Kabore KP, Fravalo P, Letellier A, and Fairbrother JM (2015) Impact of medicated feed along with clay mineral supplementation on Escherichia coli resistance to antimicrobial agents in pigs after weaning in field conditions. Res Vet Sci 102: 72-79. [Crossref]
- Vahjen W, Pietruszyńska D, Starke IC, Zentek J (2015) High dietary zinc supplementation increases the occurrence of tetracycline and sulfonamide resistance genes in the intestine of weaned pigs. Gut Pathog 7: 23. [Crossref]
- Suau A, et al. (1999) Direct Analysis of Genes Encoding 16S rRNA from Complex Communities Reveals Many Novel Molecular Species within the Human Gut. Appl Environ Microbiol 65: 4799-4807. [Crossref]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, et al. (2005) Diversity of the human intestinal microbial flora. Science 308: 1635-1638. [Crossref]
- Bryan A, Shapir N, and Sadowsky MJ (2004) Frequency and Distribution of Tetracycline Resistance Genes in Genetically Diverse, Nonselected, and Nonclinical Escherichia coli Strains Isolated from Diverse Human and Animal Sources. Appl Environ Microbiol. 70: 2503-2507. [Crossref]
- Chopra I and Roberts M (2001) Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance. Microbiol Mol Biol Rev 65: 232-260. [Crossref]
- Enne VI, Cassar C, Sprigings K, Woodward MJ, and Bennett PM (2008) A high prevalence of antimicrobial resistant Escherichia coli isolated from pigs and a low prevalence of antimicrobial resistant E. coli from cattle and sheep in Great Britain at slaughter. FEMS Microbiol Lett. 278: 193-199. [Crossref]
- Karczmarczyk M, Walsh C, Slowey R, Leonard N, and Fanning S (2011) Molecular Characterization of Multidrug-Resistant Escherichia coli Isolates from Irish Cattle Farms. Appl Environ Microbiol. 77: 7121-7127. [Crossref]
- Knapp CW, Dolfing J, Ehlert PA, and Graham DW (2010) Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environmental Science & Technology 44: 580-587.
- Agga GE, et al. (2015) Effects of chlortetracycline and copper supplementation on the prevalence, distribution, and quantity of antimicrobial resistance genes in the fecal metagenome of weaned pigs. Prev Vet Med 119: 179-189. [Crossref]
- Looft T, Johnson TA, Allen HK, Bayles DO, Alt DP, et al. (2012) In-feed antibiotic effects on the swine intestinal microbiome. Proc Natl Acad Sci U S A 109: 1691-1696. [Crossref]
- Kim HB, Borewicz K, White BA, Singer RS, Sreevatsan S, et al. (2011) Longitudinal investigation of the age-related bacterial diversity in the feces of commercial pigs. Vet Microbiol 153: 124-133. [Crossref]
- Vieira AR, Houe H, Wegener HC, Lo Fo Wong DMA, and Emborg H-D (2008) Association Between Tetracycline Consumption and Tetracycline Resistance in Escherichia coli from Healthy Danish Slaughter Pigs. Foodborne Pathog Dis. 6: 99-109. [Crossref]
- Mazurek J, Pusz P, Bok E, Stosik M, and Baldy-Chudzik K (2013) The phenotypic and genotypic characteristics of antibiotic resistance in Escherichia coli populations isolated from farm animals with different exposure to antimicrobial agents. Pol J Microbiol. 62: 173-179. [Crossref]
- Alexander TW et al. (2011) Longitudinal characterization of antimicrobial resistance genes in feces shed from cattle fed different subtherapeutic antibiotics. BMC Microbiol. 11: 19. [Crossref]
- Rosengren LB, Waldner CL, and Reid-Smith RJ (2009) Associations between Antimicrobial Resistance Phenotypes, Antimicrobial Resistance Genes, and Virulence Genes of Fecal Escherichia coli Isolates from Healthy Grow-Finish Pigs. Appl Environ Microbiol. 75: 1373-1380. [Crossref]
- Blake DP Humphry RW, Scott KP, Hillman K, Fenlon DR, et al. (2003) Influence of tetracycline exposure on tetracycline resistance and the carriage of tetracycline resistance genes within commensal Escherichia coli populations. J Appl Microbiol. 94: 1087-1097. [Crossref]
- Lee C, Langlois BE, and Dawson KA (1993) Detection of tetracycline resistance determinants in pig isolates from three herds with different histories of antimicrobial agent exposure. Appl Environ Microbiol. 59: 1467-1472. [Crossref]
- Ben-Mahrez K, Sioud M (2010) Presence of STRA-STRB linked streptomycin-resistance genes in clinical isolate of Escherichia coil 2418. Arch Inst Pasteur Tunis 87: 77-81. [Crossref]
- Clifford RJ, Milillo M, Prestwood J, Quintero R, Zurawski DV, et al. (2012) Detection of bacterial 16S rRNA and identification of four clinically important bacteria by real-time PCR. PLoS One 7: e48558. [Crossref]
- Ng LK, Martin I, Alfa M, Mulvey M (2001) Multiplex PCR for the detection of tetracycline resistant genes. Mol Cell Probes 15: 209-215. [Crossref]
- Aminov RI, Chee-Sanford JC, Garrigues N, Teferedegne B, Krapac IJ, et al. (2002) Development, Validation, and Application of PCR Primers for Detection of Tetracycline Efflux Genes of Gram-Negative Bacteria. Applied and Environmental Microbiology 68: 1786-1793. [Crossref]
- Madsen L, Aarestrup FM, Olsen JE (2000) Characterisation of streptomycin resistance determinants in Danish isolates of Salmonella Typhimurium. Vet Microbiol 75: 73-82. [Crossref]

